Gene
KWMTBOMO01032
Pre Gene Modal
BGIBMGA007376
Annotation
PREDICTED:_calpain-11-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.061 PlasmaMembrane Reliability : 1.272
Sequence
CDS
ATGAAATACAGATATGGCATCAGCAACAACATCGTACCGGTCAACGGGCGCCGCGAGGAGCCCCACTCGACTCTCTCCAGGTACTGGAGCGGCTCTGAGGGTTCAGGGCGGGTCCTTGGCGAGGGCCTGTGGGAGGACCCCGAGTTCCCCGCGACCAGCACCTCTCTGGGCGACAAGAAGTTCTCCTCGAACGTTGTTTGGATGAGACCTCATGAGCTGTGCACGAGGCCTGCATTCCGGGGCGAAGGTTTCAATGATGGAGTCACTGAAGACGAGAATGATGATGACTTTGCTGCGAGATGGAGTATAGAGGTGGGTGCCGTCGGCGACAGCAGCTTATGCTGTGCCGTTGCAGCCCTGGCGCTCACCCCTCGCCTTCTGGACCGGGTGATACCGCCGCAGACCTTCAAGGTCCACTACAATGGATCTTTTAAGTTCAATTTTTGGGTGTTCGGCGCGTGGCGCGAAGTCGCCGTTGACGACAGACTCCCGACGCGAGCTGGTCGTCTCATAACTAGTCACAACGCATTACCTGACGACTTCACCCTGCCGCTCCTGGAGAAGGCTTATGCCAAGCTATACGGTTCGTACGTAGCGCTCAGGGTGGGCAGCGTGTCGCGTGCACTCCAAGATCTTTCAGGAGGTATAGTGCAGAGTTTCTCCCTTGTCCAACAGCCGAAGGGCTTGACAGCACAAGTACTGAACTCGGCTGTTCCACGGTCAACGCTATTGGTTGCTACGGTTGCGCAGGAAAAAGAAAGTGGCTGGCGACGTCTCCGCAACGGCTTGATATCAGGTCAAGCGTATTGCGTGACAGGACTGGCCAGGGTTCGAAGTGCCAACACCGCTGATGGAGAAGGAGCACTCGTGAGAGTGAGAGCTGTCAGTGGACTTGGAGAGTGGGCAGGACCCTGGGCTAGGGGCAGTGCTGAATGGAGGGCTCTTGCAGAGGCAGACAGAGACTTGTTGGCGCGACGCGCTGACCGACCTGGGGACTTCTGGATGTCGTTCCCAGACTTCGCCCGGGCCTTCACAACACTGGTCCTGGTACACGTGGGGCCGGACGATTGGCTGGTAGAGCCTGCGTTACATGGTAAGCGACCATGGCGGGCTGTGCTTGCTAGACGTAGATGGAGACGAGGATACAACGCTGGAGGACCGCCCTCTTGCATCGAAACTACAAGCACAAATCCACAATTCCACGTACAAATACCTCGCACGGAAACCGGAGGCGCGAGGAAATGTCACGTAGTAGTCTCCGTGACCCAGCAGTATGGGACCAACAAGGCGGGGAAATTACGGGCAATAGGGTTCGCTGTGTACGAGATCCCTGCAGGAGGGGGACCGCGGAGGGGAGCTCTGAGGGCTCTGGCTGGCTTGAGGGCTTTGGACGTCACACACGAGAGCAGGGCGCGCGAAGTAGCTACGTTCTTCACGCTACCTGCCGGGCAGTATTTAGTTGTGCCACATACGAGGCGTCCGCACACAGAAGCTGCTTTCCTACTGCGCATATTTACTGACGAACAGACCGATGTGTGGGAAGTCAACGATGATAATCTGATCATTCCAAACATCGGAGCCGAGTTTCTGGACGACGACCAAAGTACATCGCCGGAGCTGCAGATGGCGATCACTAATCTGTTTGGAAAAAAGGACATCGAAGAAGTTGACGCGCGGGCGCTGCGGGGGGCGGTGCGGCGCGCGAGCGGGGGTTGGCGCGCGGCGCGGGCGGCGTGCGGCGCGCTGGTGGCGGCGCGGGACGCAGGGGCGCGGGGTGGGGCGCGCGCGCGGGACGGGCCGCCGCTAGTAGCGCGCCTGCTGGCCTGGCGGGCCGCCTACACAGGGTTAGCGCCGAGCTGCAGCGCTGCCTCCTCGTACCGGCTGCGGGCGCTGCTGTGGGCGGCGGGGGTGGCGGCCTCCAACAAGGTGCTGGAGTGTCTGGTGATGCGCTTCGCCAAAAAGAACACAATATCTTTGGATGCCTACCTTATAGCCTTGGCCAAACTACATTTAGCACATGAAAGGTTTCGATGCTTAGAAGCTAAATTAAAATCGAACCCAATATCTTTGGAAGAGATGATCCTGATGACCATTTACTCGTGA
Protein
MKYRYGISNNIVPVNGRREEPHSTLSRYWSGSEGSGRVLGEGLWEDPEFPATSTSLGDKKFSSNVVWMRPHELCTRPAFRGEGFNDGVTEDENDDDFAARWSIEVGAVGDSSLCCAVAALALTPRLLDRVIPPQTFKVHYNGSFKFNFWVFGAWREVAVDDRLPTRAGRLITSHNALPDDFTLPLLEKAYAKLYGSYVALRVGSVSRALQDLSGGIVQSFSLVQQPKGLTAQVLNSAVPRSTLLVATVAQEKESGWRRLRNGLISGQAYCVTGLARVRSANTADGEGALVRVRAVSGLGEWAGPWARGSAEWRALAEADRDLLARRADRPGDFWMSFPDFARAFTTLVLVHVGPDDWLVEPALHGKRPWRAVLARRRWRRGYNAGGPPSCIETTSTNPQFHVQIPRTETGGARKCHVVVSVTQQYGTNKAGKLRAIGFAVYEIPAGGGPRRGALRALAGLRALDVTHESRAREVATFFTLPAGQYLVVPHTRRPHTEAAFLLRIFTDEQTDVWEVNDDNLIIPNIGAEFLDDDQSTSPELQMAITNLFGKKDIEEVDARALRGAVRRASGGWRAARAACGALVAARDAGARGGARARDGPPLVARLLAWRAAYTGLAPSCSAASSYRLRALLWAAGVAASNKVLECLVMRFAKKNTISLDAYLIALAKLHLAHERFRCLEAKLKSNPISLEEMILMTIYS
Summary
Cofactor
NAD(+)
Similarity
Belongs to the peptidase C2 family.
Belongs to the NAD(P)-dependent epimerase/dehydratase family.
Belongs to the GMC oxidoreductase family.
Belongs to the NAD(P)-dependent epimerase/dehydratase family.
Belongs to the GMC oxidoreductase family.
Uniprot
A0A212FAU7
A0A1W4WVY2
A0A1Y1KIH2
D6WUX3
A0A023F4D4
A0A139WDU2
+ More
A0A1B6D1V8 A0A1W4WK38 A0A1W4WW64 A0A1V1G5B6 A0A2J7RQR3 A0A146LDH1 A0A146LCI9 A0A3Q0JH90 A0A0P5GY33 A0A067RD29 A0A0K8TBC9 A0A1Y1KHZ7 A0A194QG86 A0A2S2NV42 X1WIQ5 A0A2H8TFS3 A0A0A9WA58 A0A1D2MUU5 E0VKU5 T1JFC5 A0A0N8BDJ6 A0A0P5JQU4 A0A0P5N7Z2 A0A0P5G1J9 A0A0P5EHK3 A0A0P6ACJ0 A0A0P5QXC9 A0A0P5JEH1 A0A0P5GAB2 A0A0P5PKZ5 A0A0P6DGY0 A0A0P5RMB8 A0A0N8EE27 A0A0P5GGL2 A0A0P5IP08 A0A0N8D014 A0A0P5ZIS3 A0A0P5M887 A0A0P5T0I8 A0A0P5HYT4 A0A0P5R4N8 A0A0P5CCM5 A0A0P6E6N2 A0A0P5BHN7 A0A0N8DNK3 A0A0P5BMD5 A0A0N8BTN1 A0A0N8E8D8 A0A336LXT5 A0A0P5EYN4 A0A0P5GAX4 A0A0P6H2A3 A0A0P5TLK7 A0A162Q0Y9 A0A182V3R5 A0A182KSQ7 A0A182JKJ8 A0A0P5CZ48 A0A0P6F7V8 A0A0P5E5Y7 A0A0N8D2R4 A0A182QF23 A0A182Y1U9 A0A182PBP5 A0A182XD85 A0A182FTV1 A0A182W981 A0A2C9GQW9 Q16WJ3 A0A2M4CQD7 A0A2M4CQH0 A0A336MZB3 A0A182R5G4 B0XDG9 A0A2M3Z012 W5JMS3 A0A2M4BIV2 A0A0K8TS59 A0A1J1J1C4 A0A182SIF1 A0A2M4BK18 A0A1S4H9X2 A0A182MYH8 A0A084WHZ5 A0A182U8T2
A0A1B6D1V8 A0A1W4WK38 A0A1W4WW64 A0A1V1G5B6 A0A2J7RQR3 A0A146LDH1 A0A146LCI9 A0A3Q0JH90 A0A0P5GY33 A0A067RD29 A0A0K8TBC9 A0A1Y1KHZ7 A0A194QG86 A0A2S2NV42 X1WIQ5 A0A2H8TFS3 A0A0A9WA58 A0A1D2MUU5 E0VKU5 T1JFC5 A0A0N8BDJ6 A0A0P5JQU4 A0A0P5N7Z2 A0A0P5G1J9 A0A0P5EHK3 A0A0P6ACJ0 A0A0P5QXC9 A0A0P5JEH1 A0A0P5GAB2 A0A0P5PKZ5 A0A0P6DGY0 A0A0P5RMB8 A0A0N8EE27 A0A0P5GGL2 A0A0P5IP08 A0A0N8D014 A0A0P5ZIS3 A0A0P5M887 A0A0P5T0I8 A0A0P5HYT4 A0A0P5R4N8 A0A0P5CCM5 A0A0P6E6N2 A0A0P5BHN7 A0A0N8DNK3 A0A0P5BMD5 A0A0N8BTN1 A0A0N8E8D8 A0A336LXT5 A0A0P5EYN4 A0A0P5GAX4 A0A0P6H2A3 A0A0P5TLK7 A0A162Q0Y9 A0A182V3R5 A0A182KSQ7 A0A182JKJ8 A0A0P5CZ48 A0A0P6F7V8 A0A0P5E5Y7 A0A0N8D2R4 A0A182QF23 A0A182Y1U9 A0A182PBP5 A0A182XD85 A0A182FTV1 A0A182W981 A0A2C9GQW9 Q16WJ3 A0A2M4CQD7 A0A2M4CQH0 A0A336MZB3 A0A182R5G4 B0XDG9 A0A2M3Z012 W5JMS3 A0A2M4BIV2 A0A0K8TS59 A0A1J1J1C4 A0A182SIF1 A0A2M4BK18 A0A1S4H9X2 A0A182MYH8 A0A084WHZ5 A0A182U8T2
Pubmed
EMBL
AGBW02009415
OWR50862.1
GEZM01085289
JAV60020.1
KQ971357
EFA08352.2
+ More
GBBI01002327 JAC16385.1 KYB26110.1 GEDC01019058 GEDC01017638 JAS18240.1 JAS19660.1 FX985373 BAX07386.1 NEVH01000618 PNF43167.1 GDHC01013074 JAQ05555.1 GDHC01012698 JAQ05931.1 GDIQ01238094 JAK13631.1 KK852544 KDR21632.1 GBRD01002968 GDHC01002887 JAG62853.1 JAQ15742.1 GEZM01085288 JAV60021.1 KQ459232 KPJ02461.1 GGMR01008430 MBY21049.1 ABLF02025402 ABLF02025403 GFXV01001139 MBW12944.1 GBHO01040206 JAG03398.1 LJIJ01000523 ODM96584.1 DS235250 EEB14001.1 JH432148 GDIQ01188291 JAK63434.1 GDIQ01201094 JAK50631.1 GDIQ01146012 JAL05714.1 GDIQ01247684 JAK04041.1 GDIP01146326 GDIQ01100134 JAJ77076.1 JAL51592.1 GDIP01044498 JAM59217.1 GDIQ01117622 JAL34104.1 GDIQ01225890 JAK25835.1 GDIQ01243932 JAK07793.1 GDIQ01148268 JAL03458.1 GDIQ01086686 JAN08051.1 GDIQ01117621 JAL34105.1 GDIQ01035852 JAN58885.1 GDIQ01242687 JAK09038.1 GDIQ01235637 JAK16088.1 GDIP01082135 JAM21580.1 GDIP01044499 JAM59216.1 GDIQ01159309 JAK92416.1 GDIP01139484 JAL64230.1 GDIQ01221642 JAK30083.1 GDIQ01107773 JAL43953.1 GDIP01177344 JAJ46058.1 GDIQ01066841 JAN27896.1 GDIP01184365 JAJ39037.1 GDIP01016223 JAM87492.1 GDIP01182968 JAJ40434.1 GDIQ01146011 JAL05715.1 GDIQ01051764 JAN42973.1 UFQT01000196 SSX21519.1 GDIQ01265215 JAJ86509.1 GDIQ01265214 JAJ86510.1 GDIQ01035853 JAN58884.1 GDIP01126280 JAL77434.1 LRGB01000389 KZS19307.1 GDIP01163427 JAJ59975.1 GDIQ01053405 JAN41332.1 GDIP01147678 JAJ75724.1 GDIP01074535 JAM29180.1 AXCN02001043 APCN01003286 CH477563 EAT38965.1 GGFL01003374 MBW67552.1 GGFL01003375 MBW67553.1 UFQS01002769 UFQT01002769 SSX14636.1 SSX34033.1 DS232756 EDS45447.1 GGFM01001108 MBW21859.1 ADMH02000662 ETN65401.1 GGFJ01003772 MBW52913.1 GDAI01000620 JAI16983.1 CVRI01000066 CRL06199.1 GGFJ01004160 MBW53301.1 ATLV01023900 KE525347 KFB49839.1
GBBI01002327 JAC16385.1 KYB26110.1 GEDC01019058 GEDC01017638 JAS18240.1 JAS19660.1 FX985373 BAX07386.1 NEVH01000618 PNF43167.1 GDHC01013074 JAQ05555.1 GDHC01012698 JAQ05931.1 GDIQ01238094 JAK13631.1 KK852544 KDR21632.1 GBRD01002968 GDHC01002887 JAG62853.1 JAQ15742.1 GEZM01085288 JAV60021.1 KQ459232 KPJ02461.1 GGMR01008430 MBY21049.1 ABLF02025402 ABLF02025403 GFXV01001139 MBW12944.1 GBHO01040206 JAG03398.1 LJIJ01000523 ODM96584.1 DS235250 EEB14001.1 JH432148 GDIQ01188291 JAK63434.1 GDIQ01201094 JAK50631.1 GDIQ01146012 JAL05714.1 GDIQ01247684 JAK04041.1 GDIP01146326 GDIQ01100134 JAJ77076.1 JAL51592.1 GDIP01044498 JAM59217.1 GDIQ01117622 JAL34104.1 GDIQ01225890 JAK25835.1 GDIQ01243932 JAK07793.1 GDIQ01148268 JAL03458.1 GDIQ01086686 JAN08051.1 GDIQ01117621 JAL34105.1 GDIQ01035852 JAN58885.1 GDIQ01242687 JAK09038.1 GDIQ01235637 JAK16088.1 GDIP01082135 JAM21580.1 GDIP01044499 JAM59216.1 GDIQ01159309 JAK92416.1 GDIP01139484 JAL64230.1 GDIQ01221642 JAK30083.1 GDIQ01107773 JAL43953.1 GDIP01177344 JAJ46058.1 GDIQ01066841 JAN27896.1 GDIP01184365 JAJ39037.1 GDIP01016223 JAM87492.1 GDIP01182968 JAJ40434.1 GDIQ01146011 JAL05715.1 GDIQ01051764 JAN42973.1 UFQT01000196 SSX21519.1 GDIQ01265215 JAJ86509.1 GDIQ01265214 JAJ86510.1 GDIQ01035853 JAN58884.1 GDIP01126280 JAL77434.1 LRGB01000389 KZS19307.1 GDIP01163427 JAJ59975.1 GDIQ01053405 JAN41332.1 GDIP01147678 JAJ75724.1 GDIP01074535 JAM29180.1 AXCN02001043 APCN01003286 CH477563 EAT38965.1 GGFL01003374 MBW67552.1 GGFL01003375 MBW67553.1 UFQS01002769 UFQT01002769 SSX14636.1 SSX34033.1 DS232756 EDS45447.1 GGFM01001108 MBW21859.1 ADMH02000662 ETN65401.1 GGFJ01003772 MBW52913.1 GDAI01000620 JAI16983.1 CVRI01000066 CRL06199.1 GGFJ01004160 MBW53301.1 ATLV01023900 KE525347 KFB49839.1
Proteomes
UP000007151
UP000192223
UP000007266
UP000235965
UP000079169
UP000027135
+ More
UP000053268 UP000007819 UP000094527 UP000009046 UP000076858 UP000075903 UP000075882 UP000075880 UP000075886 UP000076408 UP000075885 UP000076407 UP000069272 UP000075920 UP000075840 UP000008820 UP000075900 UP000002320 UP000000673 UP000183832 UP000075901 UP000075884 UP000030765 UP000075902
UP000053268 UP000007819 UP000094527 UP000009046 UP000076858 UP000075903 UP000075882 UP000075880 UP000075886 UP000076408 UP000075885 UP000076407 UP000069272 UP000075920 UP000075840 UP000008820 UP000075900 UP000002320 UP000000673 UP000183832 UP000075901 UP000075884 UP000030765 UP000075902
Pfam
Interpro
IPR036213
Calpain_III_sf
+ More
IPR022682 Calpain_domain_III
IPR001300 Peptidase_C2_calpain_cat
IPR022683 Calpain_III
IPR022684 Calpain_cysteine_protease
IPR038765 Papain-like_cys_pep_sf
IPR011992 EF-hand-dom_pair
IPR000169 Pept_cys_AS
IPR027424 Glucose_Oxidase_domain_2
IPR036188 FAD/NAD-bd_sf
IPR007867 GMC_OxRtase_C
IPR000172 GMC_OxRdtase_N
IPR005886 UDP_G4E
IPR036291 NAD(P)-bd_dom_sf
IPR016040 NAD(P)-bd_dom
IPR022682 Calpain_domain_III
IPR001300 Peptidase_C2_calpain_cat
IPR022683 Calpain_III
IPR022684 Calpain_cysteine_protease
IPR038765 Papain-like_cys_pep_sf
IPR011992 EF-hand-dom_pair
IPR000169 Pept_cys_AS
IPR027424 Glucose_Oxidase_domain_2
IPR036188 FAD/NAD-bd_sf
IPR007867 GMC_OxRtase_C
IPR000172 GMC_OxRdtase_N
IPR005886 UDP_G4E
IPR036291 NAD(P)-bd_dom_sf
IPR016040 NAD(P)-bd_dom
SUPFAM
Gene 3D
ProteinModelPortal
A0A212FAU7
A0A1W4WVY2
A0A1Y1KIH2
D6WUX3
A0A023F4D4
A0A139WDU2
+ More
A0A1B6D1V8 A0A1W4WK38 A0A1W4WW64 A0A1V1G5B6 A0A2J7RQR3 A0A146LDH1 A0A146LCI9 A0A3Q0JH90 A0A0P5GY33 A0A067RD29 A0A0K8TBC9 A0A1Y1KHZ7 A0A194QG86 A0A2S2NV42 X1WIQ5 A0A2H8TFS3 A0A0A9WA58 A0A1D2MUU5 E0VKU5 T1JFC5 A0A0N8BDJ6 A0A0P5JQU4 A0A0P5N7Z2 A0A0P5G1J9 A0A0P5EHK3 A0A0P6ACJ0 A0A0P5QXC9 A0A0P5JEH1 A0A0P5GAB2 A0A0P5PKZ5 A0A0P6DGY0 A0A0P5RMB8 A0A0N8EE27 A0A0P5GGL2 A0A0P5IP08 A0A0N8D014 A0A0P5ZIS3 A0A0P5M887 A0A0P5T0I8 A0A0P5HYT4 A0A0P5R4N8 A0A0P5CCM5 A0A0P6E6N2 A0A0P5BHN7 A0A0N8DNK3 A0A0P5BMD5 A0A0N8BTN1 A0A0N8E8D8 A0A336LXT5 A0A0P5EYN4 A0A0P5GAX4 A0A0P6H2A3 A0A0P5TLK7 A0A162Q0Y9 A0A182V3R5 A0A182KSQ7 A0A182JKJ8 A0A0P5CZ48 A0A0P6F7V8 A0A0P5E5Y7 A0A0N8D2R4 A0A182QF23 A0A182Y1U9 A0A182PBP5 A0A182XD85 A0A182FTV1 A0A182W981 A0A2C9GQW9 Q16WJ3 A0A2M4CQD7 A0A2M4CQH0 A0A336MZB3 A0A182R5G4 B0XDG9 A0A2M3Z012 W5JMS3 A0A2M4BIV2 A0A0K8TS59 A0A1J1J1C4 A0A182SIF1 A0A2M4BK18 A0A1S4H9X2 A0A182MYH8 A0A084WHZ5 A0A182U8T2
A0A1B6D1V8 A0A1W4WK38 A0A1W4WW64 A0A1V1G5B6 A0A2J7RQR3 A0A146LDH1 A0A146LCI9 A0A3Q0JH90 A0A0P5GY33 A0A067RD29 A0A0K8TBC9 A0A1Y1KHZ7 A0A194QG86 A0A2S2NV42 X1WIQ5 A0A2H8TFS3 A0A0A9WA58 A0A1D2MUU5 E0VKU5 T1JFC5 A0A0N8BDJ6 A0A0P5JQU4 A0A0P5N7Z2 A0A0P5G1J9 A0A0P5EHK3 A0A0P6ACJ0 A0A0P5QXC9 A0A0P5JEH1 A0A0P5GAB2 A0A0P5PKZ5 A0A0P6DGY0 A0A0P5RMB8 A0A0N8EE27 A0A0P5GGL2 A0A0P5IP08 A0A0N8D014 A0A0P5ZIS3 A0A0P5M887 A0A0P5T0I8 A0A0P5HYT4 A0A0P5R4N8 A0A0P5CCM5 A0A0P6E6N2 A0A0P5BHN7 A0A0N8DNK3 A0A0P5BMD5 A0A0N8BTN1 A0A0N8E8D8 A0A336LXT5 A0A0P5EYN4 A0A0P5GAX4 A0A0P6H2A3 A0A0P5TLK7 A0A162Q0Y9 A0A182V3R5 A0A182KSQ7 A0A182JKJ8 A0A0P5CZ48 A0A0P6F7V8 A0A0P5E5Y7 A0A0N8D2R4 A0A182QF23 A0A182Y1U9 A0A182PBP5 A0A182XD85 A0A182FTV1 A0A182W981 A0A2C9GQW9 Q16WJ3 A0A2M4CQD7 A0A2M4CQH0 A0A336MZB3 A0A182R5G4 B0XDG9 A0A2M3Z012 W5JMS3 A0A2M4BIV2 A0A0K8TS59 A0A1J1J1C4 A0A182SIF1 A0A2M4BK18 A0A1S4H9X2 A0A182MYH8 A0A084WHZ5 A0A182U8T2
PDB
1QXP
E-value=4.00071e-55,
Score=546
Ontologies
GO
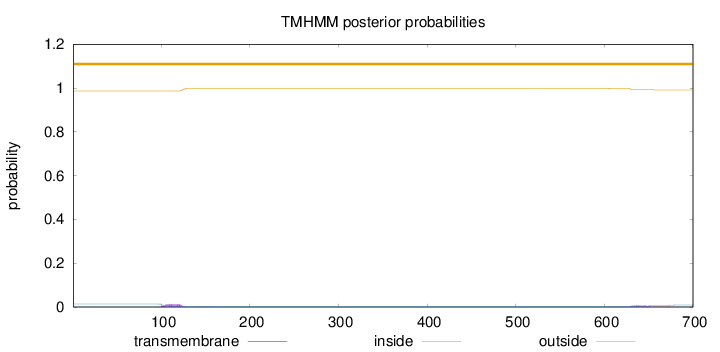
Topology
Length:
700
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43183
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01346
outside
1 - 700
Population Genetic Test Statistics
Pi
229.326209
Theta
161.816339
Tajima's D
0.888056
CLR
0.399685
CSRT
0.626518674066297
Interpretation
Uncertain
