Gene
KWMTBOMO01031
Pre Gene Modal
BGIBMGA007391
Annotation
PREDICTED:_kynurenine/alpha-aminoadipate_aminotransferase?_mitochondrial_isoform_X2_[Bombyx_mori]
Full name
Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial
Alternative Name
2-aminoadipate aminotransferase
2-aminoadipate transaminase
Alpha-aminoadipate aminotransferase
Kynurenine aminotransferase II
Kynurenine--oxoglutarate aminotransferase II
Kynurenine--oxoglutarate transaminase 2
Kynurenine--oxoglutarate transaminase II
2-aminoadipate transaminase
Alpha-aminoadipate aminotransferase
Kynurenine aminotransferase II
Kynurenine--oxoglutarate aminotransferase II
Kynurenine--oxoglutarate transaminase 2
Kynurenine--oxoglutarate transaminase II
Location in the cell
Cytoplasmic Reliability : 2.171
Sequence
CDS
ATGAAATTGACAAAGAATAATTTGGATTTAATGAAAAAAGTCTTCGAAATAAAACCTTGTGTTGTGTTGAGAGCAAAATATTCACAAGCTTGTACGCCGGAGTTCTTTAAGTTTTACGATGAGAATATTAACGTGGGTGACCATAAAGTGTTGGATGTCGAAGACTATGGTCGATTTATAAGCACCAGGTCTTCAAGGAGAGAACCAGCTATCACAAGAGTGATCACTTCCCTCGCATATAAAGTAGATAAAAAGATGATCTCCCTAGCCGAGGGTATGCCCAACGAAGGGGTGTTTCCGTTCAAGAAACTAAGCATGGACATGAAAAAGGGCGGTGGTATAATGATGGAGGGCAAAGAGCTGGCTGCGGCGTTGCAGTATGTGCCTTCACAGGGGCTTCCATCGCTACTGACGGAGCTTCGCAAATTTCAAGAGGACATACACCGTCCTCCAGACGTGCCCCGAGACCTCATGATCACCAACGGCGGGCAGCACGGCATCTACCAGTGCGCTGAGCTACTCCTGGAGCCCGGCGACCCCATCGTCACCACTGAGTACGCCTACACTGGTCTGCACAGCACCTTGAAGCCATACCAACCAGAAATCCTTGGTGTACCCGAAGACAATGACGGTCTCATCCCTGAAACACTTGAATGCATTTTGGAAGACCGACTCTCGAGGGGTCTAAAGATGCCGAAGATGATGTACGTCATTCCAACAGGCAGCAATCCTACCGGAACTGTCATCCCGGTGGAAAGGAGAATGAAAATATATGCGTTAGCTTGCAAATACGACTTTATTATTGTTGAAGATGATCCTTATATGTTCTTGAATTACAGCGGGCGTCCAATAGCTTCTTTCCTATCAATGGACAAAGCCGGTCGTGTGATCCGCCTGGATTCAGTCTCCAAGGTCGTGAGCGCTGGTCTCCGGGGAGGATGGGTAACCGCTCCATCTCCGCTATTGGAAAGACTGCAGTTGCACTCTCAAGCAGAGCTACTCCACCCATGTACCCTGTCCCAGGCGATTCTACTGAAGCTGGTTCAGTGTAGGGAGACGTTAGGTTCATATCTGAAGTCTGCGCGGGACTTGTACTGGAGCAGACGGGACGCGCTTCATTCAGCGCTGCAGCCGCTGGAGGGGCTGGCGGACTGGGCCGTTCCTGACGCTGGACTATTCTTCTGGCTGAGGGTTCGAGATGTCGACGACGTTTATAGTATGATCCACACAGCCTTCGAACGAGGTCTGATGCTGGTTCCAGGACAAGCGTTCATATACGACAGCAACGCGTCGTGCCAGTACATCAGACTGACGTTTAGCAAGATCCGACTGGAAGACATGAGCACGGCATCCCAGATATTATCGGAAGTTATAAGAGATGAACAGAAAAAGTCTTTGGAGAAGGAGCCCAAACGATACGCTACTGAGAGCTAG
Protein
MKLTKNNLDLMKKVFEIKPCVVLRAKYSQACTPEFFKFYDENINVGDHKVLDVEDYGRFISTRSSRREPAITRVITSLAYKVDKKMISLAEGMPNEGVFPFKKLSMDMKKGGGIMMEGKELAAALQYVPSQGLPSLLTELRKFQEDIHRPPDVPRDLMITNGGQHGIYQCAELLLEPGDPIVTTEYAYTGLHSTLKPYQPEILGVPEDNDGLIPETLECILEDRLSRGLKMPKMMYVIPTGSNPTGTVIPVERRMKIYALACKYDFIIVEDDPYMFLNYSGRPIASFLSMDKAGRVIRLDSVSKVVSAGLRGGWVTAPSPLLERLQLHSQAELLHPCTLSQAILLKLVQCRETLGSYLKSARDLYWSRRDALHSALQPLEGLADWAVPDAGLFFWLRVRDVDDVYSMIHTAFERGLMLVPGQAFIYDSNASCQYIRLTFSKIRLEDMSTASQILSEVIRDEQKKSLEKEPKRYATES
Summary
Description
Transaminase with broad substrate specificity. Has transaminase activity towards aminoadipate, kynurenine, methionine and glutamate. Shows activity also towards tryptophan, aspartate and hydroxykynurenine. Accepts a variety of oxo-acids as amino-group acceptors, with a preference for 2-oxoglutarate, 2-oxocaproic acid, phenylpyruvate and alpha-oxo-gamma-methiol butyric acid. Can also use glyoxylate as amino-group acceptor (in vitro).
Transaminase with broad substrate specificity. Has transaminase activity towards aminoadipate, kynurenine, methionine and glutamate. Shows activity also towards tryptophan, aspartate and hydroxykynurenine. Accepts a variety of oxo-acids as amino-group acceptors, with a preference for 2-oxoglutarate, 2-oxocaproic acid, phenylpyruvate and alpha-oxo-gamma-methiol butyric acid. Can also use glyoxylate as amino-group acceptor (in vitro) (By similarity).
Transaminase with broad substrate specificity. Has transaminase activity towards aminoadipate, kynurenine, methionine and glutamate. Shows activity also towards tryptophan, aspartate and hydroxykynurenine. Accepts a variety of oxo-acids as amino-group acceptors, with a preference for 2-oxoglutarate, 2-oxocaproic acid, phenylpyruvate and alpha-oxo-gamma-methiol butyric acid. Can also use glyoxylate as amino-group acceptor (in vitro) (By similarity).
Catalytic Activity
2-oxoglutarate + L-kynurenine = 4-(2-aminophenyl)-2,4-dioxobutanoate + L-glutamate
2-oxoglutarate + L-2-aminoadipate = 2-oxoadipate + L-glutamate
2-oxoglutarate + L-2-aminoadipate = 2-oxoadipate + L-glutamate
Cofactor
pyridoxal 5'-phosphate
Biophysicochemical Properties
0.9 mM for aminoadipate
4.7 mM for kynurenine
1.7 mM for methionine
1.6 mM for glutamate
1.8 mM for tyrosine
1.2 mM for 2-oxoglutarate
1.5 mM for 2-oxocaproic acid
1.8 mM for phenylpyruvate
1.4 mM for ino-3-pyruvate
4.7 mM for kynurenine
1.7 mM for methionine
1.6 mM for glutamate
1.8 mM for tyrosine
1.2 mM for 2-oxoglutarate
1.5 mM for 2-oxocaproic acid
1.8 mM for phenylpyruvate
1.4 mM for ino-3-pyruvate
Subunit
Homodimer.
Similarity
Belongs to the class-I pyridoxal-phosphate-dependent aminotransferase family.
Keywords
3D-structure
Acetylation
Alternative splicing
Aminotransferase
Complete proteome
Mitochondrion
Polymorphism
Pyridoxal phosphate
Reference proteome
Transferase
Transit peptide
Phosphoprotein
Direct protein sequencing
Feature
chain Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial
splice variant In isoform 2.
sequence variant In dbSNP:rs56350236.
splice variant In isoform 2.
sequence variant In dbSNP:rs56350236.
Uniprot
H9JCZ4
A0A2A4K0Z8
A0A2W1BTY4
A0A2H1WNW2
A0A194PZ24
A0A212FIT5
+ More
A0A0N1IH15 A0A212FHC4 A0A212FIU3 A0A1Y1N1Q5 A0A1W4WAJ6 D6WQD2 A0A0L7RC49 J3JTB2 A0A026WS89 A0A088AJ75 E2AUG1 A0A2A3ETA7 A0A154NX47 A0A158P0N9 E9IYK4 E2C914 A0A310S5U3 K7IWD8 A0A0J7L7M5 A0A232ELB4 F4X7L5 A0A195FXA8 A0A195B1W4 A0A0C9RU72 A0A2K6AVD3 A0A2K5U1I3 A0A0D9S243 H9EWA2 A0A2K6PXP6 A0A2K5J0A5 G3R8S4 A0A2I3H6R9 A0A2J8R340 A0A2K5XBB7 A0A2K5KS36 H2QQF3 Q4W5N8 Q8N5Z0 A0A2R9C4D7 I3NB42 A0A2K5J0L0 G7P6J8 G7MSB1 F7I8M7 C1BQK8 Q8N5Z0-2 G1R4G5 A0A2J8R368 A0A2J8MQU2 T1JA88 A0A2K5XB43 A0A195CYV6 A0A2K6T761 A0A2K5R432 A0A2Y9E9G4 L8IRI7 A0A096MU40 Q5E9N4 F6ZME2 A0A195DXF7 A0A224XNE8 F1MKS7 Q9WVM8 E2QUN4 A0A2J8R332 A0A2K6MPY3 F6XT01 A0A0P4VMR9 R4G5N9 A0A151WVV0 H0XUV2 G5BC26 A0A0P4WC97 A0A1U7QUV5 G3T487 M3WH83 A0A2Y9KIP4 A0A1B6EA68 L5LGX7 A0A2K5C1G8 E0V9A9 A0A1B6DTV0 A0A384BNL8 S7P2G6 A0A069DTV3 Q64602 A0A2U3VE14 A0A023F7S7 A0A1D2NBY8 A0A2Y9T8G1 A0A1L8HLG6 A0A0M8ZTE7 A0A1U7U0N8
A0A0N1IH15 A0A212FHC4 A0A212FIU3 A0A1Y1N1Q5 A0A1W4WAJ6 D6WQD2 A0A0L7RC49 J3JTB2 A0A026WS89 A0A088AJ75 E2AUG1 A0A2A3ETA7 A0A154NX47 A0A158P0N9 E9IYK4 E2C914 A0A310S5U3 K7IWD8 A0A0J7L7M5 A0A232ELB4 F4X7L5 A0A195FXA8 A0A195B1W4 A0A0C9RU72 A0A2K6AVD3 A0A2K5U1I3 A0A0D9S243 H9EWA2 A0A2K6PXP6 A0A2K5J0A5 G3R8S4 A0A2I3H6R9 A0A2J8R340 A0A2K5XBB7 A0A2K5KS36 H2QQF3 Q4W5N8 Q8N5Z0 A0A2R9C4D7 I3NB42 A0A2K5J0L0 G7P6J8 G7MSB1 F7I8M7 C1BQK8 Q8N5Z0-2 G1R4G5 A0A2J8R368 A0A2J8MQU2 T1JA88 A0A2K5XB43 A0A195CYV6 A0A2K6T761 A0A2K5R432 A0A2Y9E9G4 L8IRI7 A0A096MU40 Q5E9N4 F6ZME2 A0A195DXF7 A0A224XNE8 F1MKS7 Q9WVM8 E2QUN4 A0A2J8R332 A0A2K6MPY3 F6XT01 A0A0P4VMR9 R4G5N9 A0A151WVV0 H0XUV2 G5BC26 A0A0P4WC97 A0A1U7QUV5 G3T487 M3WH83 A0A2Y9KIP4 A0A1B6EA68 L5LGX7 A0A2K5C1G8 E0V9A9 A0A1B6DTV0 A0A384BNL8 S7P2G6 A0A069DTV3 Q64602 A0A2U3VE14 A0A023F7S7 A0A1D2NBY8 A0A2Y9T8G1 A0A1L8HLG6 A0A0M8ZTE7 A0A1U7U0N8
Pubmed
19121390
28756777
26354079
22118469
28004739
18362917
+ More
19820115 22516182 23537049 24508170 30249741 20798317 21347285 21282665 20075255 28648823 21719571 17431167 25319552 25362486 22398555 16136131 11181995 12126930 14702039 15489334 24275569 18620547 18056995 18056996 22722832 22002653 25243066 22751099 16305752 19892987 19393038 10441733 16141072 21183079 23806337 23576753 16341006 18464734 27129103 21993625 17975172 20566863 24813606 26334808 7493966 22673903 25474469 27289101 27762356
19820115 22516182 23537049 24508170 30249741 20798317 21347285 21282665 20075255 28648823 21719571 17431167 25319552 25362486 22398555 16136131 11181995 12126930 14702039 15489334 24275569 18620547 18056995 18056996 22722832 22002653 25243066 22751099 16305752 19892987 19393038 10441733 16141072 21183079 23806337 23576753 16341006 18464734 27129103 21993625 17975172 20566863 24813606 26334808 7493966 22673903 25474469 27289101 27762356
EMBL
BABH01027601
BABH01027602
NWSH01000308
PCG77564.1
KZ149970
PZC76096.1
+ More
ODYU01009922 SOQ54666.1 KQ459583 KPI98582.1 AGBW02008333 OWR53639.1 KQ459764 KPJ20110.1 AGBW02008507 OWR53134.1 OWR53641.1 GEZM01019993 GEZM01019992 JAV89477.1 KQ971354 EFA06089.1 KQ414617 KOC68398.1 APGK01024456 APGK01024457 APGK01024458 BT126468 KB740546 AEE61432.1 ENN80403.1 KK107138 QOIP01000002 EZA57959.1 RLU25562.1 GL442820 EFN62920.1 KZ288185 PBC34985.1 KQ434777 KZC04173.1 ADTU01005655 GL767018 EFZ14351.1 GL453765 EFN75570.1 KQ769255 OAD52887.1 LBMM01000371 KMQ99043.1 NNAY01003580 OXU19156.1 GL888862 EGI57544.1 KQ981200 KYN45061.1 KQ976667 KYM78277.1 GBYB01012290 JAG82057.1 AQIA01057100 AQIA01057101 AQIB01017787 JSUE03034993 JU322905 JU472693 JU472694 JU472695 JU472696 JU472697 AFE66661.1 AFH29497.1 CABD030034902 ADFV01179160 ADFV01179161 ABGA01096877 ABGA01096878 NDHI03003780 PNJ02939.1 PNJ02941.1 PNJ02943.1 AACZ04005183 GABC01008201 GABF01008718 GABF01008717 GABD01010378 GABE01007331 NBAG03000247 JAA03137.1 JAA13427.1 JAA22722.1 JAA37408.1 PNI61883.1 PNI61884.1 PNI61885.1 AC084866 AK314729 CH471056 AAY41030.1 BAG37271.1 EAX04771.1 AF097994 AF481738 AK055952 BC031068 AJFE02075193 AJFE02075194 AGTP01039447 AGTP01039448 CM001280 EHH54066.1 CM001257 EHH26305.1 GAMT01007577 GAMS01008816 GAMR01005060 GAMR01005059 GAMR01005058 GAMQ01006989 GAMP01000032 JAB04284.1 JAB14320.1 JAB28872.1 JAB34862.1 JAB52723.1 BT076887 ACO11311.1 PNJ02942.1 PNI61882.1 JH431987 KQ977141 KYN05324.1 JH880814 ELR58668.1 AHZZ02028238 BT020844 BT020886 BC109974 KQ980199 KYN17294.1 GFTR01006466 JAW09960.1 AF072376 AK075578 BC012637 AAEX03014301 PNJ02944.1 AAPN01147876 AAPN01147877 AAPN01147878 AAPN01147879 GDKW01002836 JAI53759.1 GAHY01000365 JAA77145.1 KQ982691 KYQ52013.1 AAQR03016671 JH169448 GEBF01007365 EHB06837.1 JAN96267.1 GDRN01064415 JAI64842.1 AANG04001605 GEDC01002491 JAS34807.1 KB112375 ELK24903.1 DS234991 EEB09965.1 GEDC01008229 JAS29069.1 KE161520 EPQ04323.1 GBGD01001371 JAC87518.1 Z50144 BC078864 GBBI01001417 JAC17295.1 LJIJ01000104 ODN02595.1 CM004467 OCT96944.1 KQ435851 KOX70755.1
ODYU01009922 SOQ54666.1 KQ459583 KPI98582.1 AGBW02008333 OWR53639.1 KQ459764 KPJ20110.1 AGBW02008507 OWR53134.1 OWR53641.1 GEZM01019993 GEZM01019992 JAV89477.1 KQ971354 EFA06089.1 KQ414617 KOC68398.1 APGK01024456 APGK01024457 APGK01024458 BT126468 KB740546 AEE61432.1 ENN80403.1 KK107138 QOIP01000002 EZA57959.1 RLU25562.1 GL442820 EFN62920.1 KZ288185 PBC34985.1 KQ434777 KZC04173.1 ADTU01005655 GL767018 EFZ14351.1 GL453765 EFN75570.1 KQ769255 OAD52887.1 LBMM01000371 KMQ99043.1 NNAY01003580 OXU19156.1 GL888862 EGI57544.1 KQ981200 KYN45061.1 KQ976667 KYM78277.1 GBYB01012290 JAG82057.1 AQIA01057100 AQIA01057101 AQIB01017787 JSUE03034993 JU322905 JU472693 JU472694 JU472695 JU472696 JU472697 AFE66661.1 AFH29497.1 CABD030034902 ADFV01179160 ADFV01179161 ABGA01096877 ABGA01096878 NDHI03003780 PNJ02939.1 PNJ02941.1 PNJ02943.1 AACZ04005183 GABC01008201 GABF01008718 GABF01008717 GABD01010378 GABE01007331 NBAG03000247 JAA03137.1 JAA13427.1 JAA22722.1 JAA37408.1 PNI61883.1 PNI61884.1 PNI61885.1 AC084866 AK314729 CH471056 AAY41030.1 BAG37271.1 EAX04771.1 AF097994 AF481738 AK055952 BC031068 AJFE02075193 AJFE02075194 AGTP01039447 AGTP01039448 CM001280 EHH54066.1 CM001257 EHH26305.1 GAMT01007577 GAMS01008816 GAMR01005060 GAMR01005059 GAMR01005058 GAMQ01006989 GAMP01000032 JAB04284.1 JAB14320.1 JAB28872.1 JAB34862.1 JAB52723.1 BT076887 ACO11311.1 PNJ02942.1 PNI61882.1 JH431987 KQ977141 KYN05324.1 JH880814 ELR58668.1 AHZZ02028238 BT020844 BT020886 BC109974 KQ980199 KYN17294.1 GFTR01006466 JAW09960.1 AF072376 AK075578 BC012637 AAEX03014301 PNJ02944.1 AAPN01147876 AAPN01147877 AAPN01147878 AAPN01147879 GDKW01002836 JAI53759.1 GAHY01000365 JAA77145.1 KQ982691 KYQ52013.1 AAQR03016671 JH169448 GEBF01007365 EHB06837.1 JAN96267.1 GDRN01064415 JAI64842.1 AANG04001605 GEDC01002491 JAS34807.1 KB112375 ELK24903.1 DS234991 EEB09965.1 GEDC01008229 JAS29069.1 KE161520 EPQ04323.1 GBGD01001371 JAC87518.1 Z50144 BC078864 GBBI01001417 JAC17295.1 LJIJ01000104 ODN02595.1 CM004467 OCT96944.1 KQ435851 KOX70755.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000192223
+ More
UP000007266 UP000053825 UP000019118 UP000053097 UP000279307 UP000005203 UP000000311 UP000242457 UP000076502 UP000005205 UP000008237 UP000002358 UP000036403 UP000215335 UP000007755 UP000078541 UP000078540 UP000233120 UP000233100 UP000029965 UP000006718 UP000233200 UP000233080 UP000001519 UP000001073 UP000001595 UP000233140 UP000233060 UP000002277 UP000005640 UP000240080 UP000005215 UP000009130 UP000008225 UP000078542 UP000233220 UP000233040 UP000248480 UP000028761 UP000009136 UP000002281 UP000078492 UP000000589 UP000002254 UP000233180 UP000002279 UP000075809 UP000005225 UP000006813 UP000189706 UP000007646 UP000011712 UP000248482 UP000233020 UP000009046 UP000261680 UP000291021 UP000002494 UP000245340 UP000094527 UP000248484 UP000186698 UP000053105 UP000189704
UP000007266 UP000053825 UP000019118 UP000053097 UP000279307 UP000005203 UP000000311 UP000242457 UP000076502 UP000005205 UP000008237 UP000002358 UP000036403 UP000215335 UP000007755 UP000078541 UP000078540 UP000233120 UP000233100 UP000029965 UP000006718 UP000233200 UP000233080 UP000001519 UP000001073 UP000001595 UP000233140 UP000233060 UP000002277 UP000005640 UP000240080 UP000005215 UP000009130 UP000008225 UP000078542 UP000233220 UP000233040 UP000248480 UP000028761 UP000009136 UP000002281 UP000078492 UP000000589 UP000002254 UP000233180 UP000002279 UP000075809 UP000005225 UP000006813 UP000189706 UP000007646 UP000011712 UP000248482 UP000233020 UP000009046 UP000261680 UP000291021 UP000002494 UP000245340 UP000094527 UP000248484 UP000186698 UP000053105 UP000189704
Interpro
Gene 3D
ProteinModelPortal
H9JCZ4
A0A2A4K0Z8
A0A2W1BTY4
A0A2H1WNW2
A0A194PZ24
A0A212FIT5
+ More
A0A0N1IH15 A0A212FHC4 A0A212FIU3 A0A1Y1N1Q5 A0A1W4WAJ6 D6WQD2 A0A0L7RC49 J3JTB2 A0A026WS89 A0A088AJ75 E2AUG1 A0A2A3ETA7 A0A154NX47 A0A158P0N9 E9IYK4 E2C914 A0A310S5U3 K7IWD8 A0A0J7L7M5 A0A232ELB4 F4X7L5 A0A195FXA8 A0A195B1W4 A0A0C9RU72 A0A2K6AVD3 A0A2K5U1I3 A0A0D9S243 H9EWA2 A0A2K6PXP6 A0A2K5J0A5 G3R8S4 A0A2I3H6R9 A0A2J8R340 A0A2K5XBB7 A0A2K5KS36 H2QQF3 Q4W5N8 Q8N5Z0 A0A2R9C4D7 I3NB42 A0A2K5J0L0 G7P6J8 G7MSB1 F7I8M7 C1BQK8 Q8N5Z0-2 G1R4G5 A0A2J8R368 A0A2J8MQU2 T1JA88 A0A2K5XB43 A0A195CYV6 A0A2K6T761 A0A2K5R432 A0A2Y9E9G4 L8IRI7 A0A096MU40 Q5E9N4 F6ZME2 A0A195DXF7 A0A224XNE8 F1MKS7 Q9WVM8 E2QUN4 A0A2J8R332 A0A2K6MPY3 F6XT01 A0A0P4VMR9 R4G5N9 A0A151WVV0 H0XUV2 G5BC26 A0A0P4WC97 A0A1U7QUV5 G3T487 M3WH83 A0A2Y9KIP4 A0A1B6EA68 L5LGX7 A0A2K5C1G8 E0V9A9 A0A1B6DTV0 A0A384BNL8 S7P2G6 A0A069DTV3 Q64602 A0A2U3VE14 A0A023F7S7 A0A1D2NBY8 A0A2Y9T8G1 A0A1L8HLG6 A0A0M8ZTE7 A0A1U7U0N8
A0A0N1IH15 A0A212FHC4 A0A212FIU3 A0A1Y1N1Q5 A0A1W4WAJ6 D6WQD2 A0A0L7RC49 J3JTB2 A0A026WS89 A0A088AJ75 E2AUG1 A0A2A3ETA7 A0A154NX47 A0A158P0N9 E9IYK4 E2C914 A0A310S5U3 K7IWD8 A0A0J7L7M5 A0A232ELB4 F4X7L5 A0A195FXA8 A0A195B1W4 A0A0C9RU72 A0A2K6AVD3 A0A2K5U1I3 A0A0D9S243 H9EWA2 A0A2K6PXP6 A0A2K5J0A5 G3R8S4 A0A2I3H6R9 A0A2J8R340 A0A2K5XBB7 A0A2K5KS36 H2QQF3 Q4W5N8 Q8N5Z0 A0A2R9C4D7 I3NB42 A0A2K5J0L0 G7P6J8 G7MSB1 F7I8M7 C1BQK8 Q8N5Z0-2 G1R4G5 A0A2J8R368 A0A2J8MQU2 T1JA88 A0A2K5XB43 A0A195CYV6 A0A2K6T761 A0A2K5R432 A0A2Y9E9G4 L8IRI7 A0A096MU40 Q5E9N4 F6ZME2 A0A195DXF7 A0A224XNE8 F1MKS7 Q9WVM8 E2QUN4 A0A2J8R332 A0A2K6MPY3 F6XT01 A0A0P4VMR9 R4G5N9 A0A151WVV0 H0XUV2 G5BC26 A0A0P4WC97 A0A1U7QUV5 G3T487 M3WH83 A0A2Y9KIP4 A0A1B6EA68 L5LGX7 A0A2K5C1G8 E0V9A9 A0A1B6DTV0 A0A384BNL8 S7P2G6 A0A069DTV3 Q64602 A0A2U3VE14 A0A023F7S7 A0A1D2NBY8 A0A2Y9T8G1 A0A1L8HLG6 A0A0M8ZTE7 A0A1U7U0N8
PDB
2VGZ
E-value=3.68148e-85,
Score=803
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion
Nucleus
Nucleus
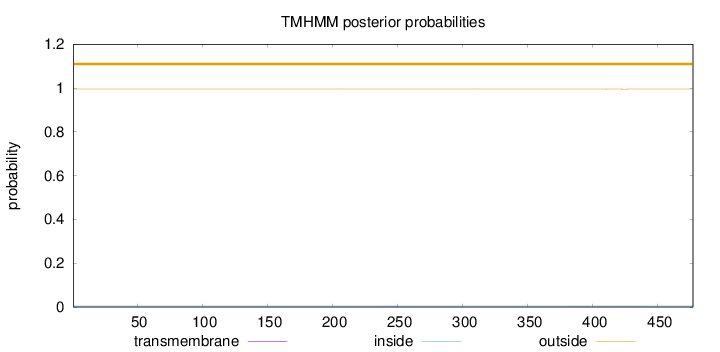
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02123
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00438
outside
1 - 477
Population Genetic Test Statistics
Pi
188.878295
Theta
203.085069
Tajima's D
0.418138
CLR
0.309025
CSRT
0.491175441227939
Interpretation
Uncertain
