Gene
KWMTBOMO01030 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007390
Annotation
PREDICTED:_microsomal_triglyceride_transfer_protein_large_subunit-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.174
Sequence
CDS
ATGTTAGTTTTAAGTGTACTGATAGCTTTTTACGTGTTTAGTGATTTTTTTTATATTACGAACGCAATACCGAAGAATGAGCATGAAATAAAACTGTTTCAAACTAGTTACAATTTTGATGTTGATGTGACGGTTTTATTAAATGATGTTAACCGAACAGATAAAGAGATAGGATACAAGATAAAGGCGACCCTTGGGGTCACGCCTGTGTGGAAGAGTGATGACTCCGAGTTCCTTTTAAAATTTGATTTACGCAATCCAAAACTACATTTAAGAGGAAAGCATCCAACAGCTGAGTTCCTACCGCGCCCTTCACCCTTAGACTCGTACTCAGAGGCACCATTTTACGCATTATGGAACAAAGGAACAGTCGAAACTTTATATTTAGATCCAAAGGAAATGAAAGATGTATACAATTTGAAGAAAGCGCTGGCTAGTCTTTTCCAGTTTCACATAATAGACGGAGAATACAAGGAGACTGACATATCAGGCGAATGTGACGTCATTTACGAAAGCATTTCGGAAAATGTGTTCAGGAAAATGAAGAGACGGTGCTCGTGGGAGTCGGCGCAGGTGGGGTGCGCGCGCGTGGCCCGCTACACGCTGCGGCCGGCGCGGGACGGACTCCTGGCCGTGTACTCCGAGGAGCTGCTGACCGTGGGCGACCACTACGGACTCAAGTTCAGGTCCTTCGTGTCCCTGAAGGCCAAAGAAGATTCCGAAGCCGCGAAACCTTCGACTGAAGAATTGGACGTGGCGTTGCACGGTCTGCCGGCGAATTTAAAACCTATGGGCTTACCGATGGTTTTAAGTGCCGACGACGAAAAGGAATACCAGACGCTGACGGAAGCAATTGACGCGAACCTAGCGGCTCTACAAGTGGAGGAGGCGGGGGCGGGCGGCACCAGCGAGGCGGCCGACGCCACGTTAAACTTACTGCCGGCGGTGAGGGCTGCTACGTTCGAAGAGTTGCGGTCGGTGCTGGAAAATGAAGAGAATCATCAACTGCTGTCGGGGCTGTGCCGGCTGGTGGGGGCGGGCGGCACGGCGGCGGGGCTGCGCGCGGTGGACGCGCTGCTGCAGCTGCGCTCGGACGACCCCGTGCTGCACCTGGCGCACATCTACCTGCAGGGGTTCGCGCTGTCCGACGCGCCGCAGGTGGATGCAGTGCGAGAGATTCTGAAGATAGGTGAGAACGGTAAAACTCGCTCCGTGGCCGAGAGCGCGCTGCTGGCCGCCGCTGCAGCAGCACAGACCCTCGCCGCCGCCGGGGACCAGGGGCTGGCCGTCGCCGTCAAGGACTCGCTCAGTAAAGGACTTGCTAAATGTAAGGACGACGACTGCAGGACAGTGCGAGTGCAGGCCCTCGCGAACCTGCGGCGCCCCGACACCGCCGAGCTGCTGCTGCAGCACGCCGAGAGGAAGGAGTCCCCCGTGTCGCTCCGCGTGGTCGCGCTGGAGGGGCTGGGCGCCGTGCCGCTGCCGCACGACAAGCTGCACCGCCTCACGCACGTGGCGCTGGACGACGCCGCGCCCCTCGAGGTGCGCGCGGCGGCGCTGGACCTGGTGGTGCTGCGCCACGTGCACGTGCCCCTTCCGCTGGCCCGGGTCGCCTCGAAGCTGCACTTCAGGTCGCCGCCCGAGCTGCGCCGTATCTTCTGGCAGCGGTTCAGAGCCCTGGCCGAGGTGCACGACTCCATGAGGGACCTGTACGCGCGCATGGGCATCAGCTTCAAGTCCTGGGACGCGCAGGCGCTCAACGGCACGTCGTCGGTGCTGGTGCGGAACCCGGGGTGGGAGGCGGCGGGCTGGTCCAGTAAGCTGGAGTCGGTGCAGCTGGCGCAGGGCGGGCTGCTGCGGCGGGGCGTGGTGCGGCTGCTGGGCTCGCGGGGCGGAGCGCCCGACGCGGAGGCGCTGGCCGTGGAAGTGTGGACCCGGGGCCTGGAGGCGCTGGCCGGCGGGGGCGACCAGGAGCCCGAGGGGGCGAGCCGCGGGGCGGAGGAGGCGGGCATGGCGGGGGGATGGCGCTGCGGAGTTGGTATTGTGTATTGCGAGCAGGCGGCGCTGCTGGCGCACGTGTGGGCGGGCACGGCGGCGGCGCCCACGCCCGTGGCCCGCGCGCTGCTGCCTCTGGTCTCCTTGCACGCCCGCCTGCCGCTGAGCGCCGGGCTGGTGCTGAGTGTGCGACACGACGCGTTGCTGGCGCTGTCCGTGGACGCCAGCGTGCAGGTGTCGCTGTGGTGGCGCACGGCCCGCGCCGAGCTGGGGCTGCGCGCCGCCGCCGCCGCGCGGGTGCTCGCGGGGGGCGCGGCGCTGGGGGGCGCGCTGCGGGCGCGGGTGCGCGGGGGGGCGGCGCCGGCGGCCACCATCTCCTCGGACCTGGACTTCTACTCCGACGTGGCGCTGTGCGTGCGGACCAACATCCACCCTTATGAAATCAGACGCAACGTGACCCTAGACAGCTCGCAGGGCGCCAGGGGGCTGCGCGTACGGCGCTCCCGGGACAGCTCGCTGCGGCTGGCGGGCCGCACGCTGTCCCTGGGCCGCCCCAACGACCGCACGTGCGGCGCGCTGCGTGCGGCCGACGCCGAGTCCTCCTAG
Protein
MLVLSVLIAFYVFSDFFYITNAIPKNEHEIKLFQTSYNFDVDVTVLLNDVNRTDKEIGYKIKATLGVTPVWKSDDSEFLLKFDLRNPKLHLRGKHPTAEFLPRPSPLDSYSEAPFYALWNKGTVETLYLDPKEMKDVYNLKKALASLFQFHIIDGEYKETDISGECDVIYESISENVFRKMKRRCSWESAQVGCARVARYTLRPARDGLLAVYSEELLTVGDHYGLKFRSFVSLKAKEDSEAAKPSTEELDVALHGLPANLKPMGLPMVLSADDEKEYQTLTEAIDANLAALQVEEAGAGGTSEAADATLNLLPAVRAATFEELRSVLENEENHQLLSGLCRLVGAGGTAAGLRAVDALLQLRSDDPVLHLAHIYLQGFALSDAPQVDAVREILKIGENGKTRSVAESALLAAAAAAQTLAAAGDQGLAVAVKDSLSKGLAKCKDDDCRTVRVQALANLRRPDTAELLLQHAERKESPVSLRVVALEGLGAVPLPHDKLHRLTHVALDDAAPLEVRAAALDLVVLRHVHVPLPLARVASKLHFRSPPELRRIFWQRFRALAEVHDSMRDLYARMGISFKSWDAQALNGTSSVLVRNPGWEAAGWSSKLESVQLAQGGLLRRGVVRLLGSRGGAPDAEALAVEVWTRGLEALAGGGDQEPEGASRGAEEAGMAGGWRCGVGIVYCEQAALLAHVWAGTAAAPTPVARALLPLVSLHARLPLSAGLVLSVRHDALLALSVDASVQVSLWWRTARAELGLRAAAAARVLAGGAALGGALRARVRGGAAPAATISSDLDFYSDVALCVRTNIHPYEIRRNVTLDSSQGARGLRVRRSRDSSLRLAGRTLSLGRPNDRTCGALRAADAESS
Summary
Uniprot
H9JCZ3
A0A194QAK0
A0A212FAV5
A0A195CS37
E9JDB1
A0A195FTB1
+ More
A0A158NZ25 A0A195BL24 A0A026WL08 A0A151J0T2 E2A7J6 F4WFP2 A0A151WI75 A0A1Q3FP29 A0A0N0U5B8 D6WXA1 A0A0L7QN03 A0A0P5HVM8 A0A0N8ATA3 A0A0P6F6R3 A0A0P5LNU4 A0A0P5HWW3 A0A0P5J4R7 A0A0P5PEN7 A0A0P5PYN4 A0A0P5N9T1 A0A0P5NZI7 A0A0P5W1X5 A0A0P5NZF1 A0A0N8C0Y6 A0A0P5PPV8 A0A0P5Z5W1 A0A0P5I5L6 A0A0N8B815 A0A0P5PTD7 A0A0P5D8X5 A0A0P5HCD4 A0A0P5F0U4 A0A0P6IGL0 A0A0P5D4S9 A0A0P5WHD6 A0A0P5D666 A0A0N8BGW3 A0A0P5ATT2 A0A0P4YI45 A0A0P5VCJ9 A0A0P5PSY4 A0A0P5UNM9 A0A0P5DQT7 A0A0P5A513 A0A0P4ZE08 A0A0P5B8E6 A0A0P6CRP7 A0A0P5WPE5 A0A0N8BQA0 A0A0P4Z7P5 A0A1W4WNT9 A0A0P4ZUQ2 A0A0P5MSZ7 A0A0P5ARV5 A0A0N8BNT7 A0A0P5M7K6 A0A0P5VCJ7 A0A0P6DN31 A0A0N8E035 A0A0P5TDL9 A0A0P6EI51 A0A0P5V4E0 A0A0P5UQG3
A0A158NZ25 A0A195BL24 A0A026WL08 A0A151J0T2 E2A7J6 F4WFP2 A0A151WI75 A0A1Q3FP29 A0A0N0U5B8 D6WXA1 A0A0L7QN03 A0A0P5HVM8 A0A0N8ATA3 A0A0P6F6R3 A0A0P5LNU4 A0A0P5HWW3 A0A0P5J4R7 A0A0P5PEN7 A0A0P5PYN4 A0A0P5N9T1 A0A0P5NZI7 A0A0P5W1X5 A0A0P5NZF1 A0A0N8C0Y6 A0A0P5PPV8 A0A0P5Z5W1 A0A0P5I5L6 A0A0N8B815 A0A0P5PTD7 A0A0P5D8X5 A0A0P5HCD4 A0A0P5F0U4 A0A0P6IGL0 A0A0P5D4S9 A0A0P5WHD6 A0A0P5D666 A0A0N8BGW3 A0A0P5ATT2 A0A0P4YI45 A0A0P5VCJ9 A0A0P5PSY4 A0A0P5UNM9 A0A0P5DQT7 A0A0P5A513 A0A0P4ZE08 A0A0P5B8E6 A0A0P6CRP7 A0A0P5WPE5 A0A0N8BQA0 A0A0P4Z7P5 A0A1W4WNT9 A0A0P4ZUQ2 A0A0P5MSZ7 A0A0P5ARV5 A0A0N8BNT7 A0A0P5M7K6 A0A0P5VCJ7 A0A0P6DN31 A0A0N8E035 A0A0P5TDL9 A0A0P6EI51 A0A0P5V4E0 A0A0P5UQG3
Pubmed
EMBL
BABH01027596
BABH01027597
KQ459232
KPJ02459.1
AGBW02009415
OWR50861.1
+ More
KQ977329 KYN03511.1 GL771866 EFZ09266.1 KQ981276 KYN43542.1 ADTU01004530 KQ976444 KYM86227.1 KK107161 QOIP01000003 EZA56613.1 RLU24874.1 KQ980591 KYN15210.1 GL437344 EFN70589.1 GL888120 EGI67019.1 KQ983089 KYQ47538.1 GFDL01005773 JAV29272.1 KQ435782 KOX74771.1 KQ971361 EFA08014.1 KQ414864 KOC60003.1 GDIQ01245036 JAK06689.1 GDIQ01245035 JAK06690.1 GDIQ01064698 JAN30039.1 GDIQ01169413 JAK82312.1 GDIQ01226700 JAK25025.1 GDIQ01205298 JAK46427.1 GDIQ01135719 JAL16007.1 GDIQ01142316 JAL09410.1 GDIQ01145274 JAL06452.1 GDIQ01135718 JAL16008.1 GDIP01093029 JAM10686.1 GDIQ01135720 JAL16006.1 GDIQ01125571 GDIQ01052465 JAL26155.1 GDIQ01241140 GDIQ01125569 GDIQ01039403 JAL26157.1 GDIP01061184 JAM42531.1 GDIQ01241141 GDIQ01125570 GDIQ01066049 JAK10584.1 GDIQ01203739 JAK47986.1 GDIQ01123722 JAL28004.1 GDIP01159276 JAJ64126.1 GDIQ01235742 JAK15983.1 GDIP01152974 JAJ70428.1 GDIQ01004723 JAN90014.1 GDIP01160931 JAJ62471.1 GDIP01086430 JAM17285.1 GDIP01162420 JAJ60982.1 GDIQ01178955 JAK72770.1 GDIP01198080 JAJ25322.1 GDIP01227412 GDIP01194978 GDIP01143775 GDIP01124135 JAI95989.1 GDIP01101619 JAM02096.1 GDIQ01123721 JAL28005.1 GDIP01112436 JAL91278.1 GDIP01152071 JAJ71331.1 GDIP01216316 JAJ07086.1 GDIP01218308 JAJ05094.1 GDIP01203461 JAJ19941.1 GDIP01010066 JAM93649.1 GDIP01083434 JAM20281.1 GDIQ01155459 JAK96266.1 GDIP01218307 JAJ05095.1 GDIP01209368 JAJ14034.1 GDIQ01159564 JAK92161.1 GDIP01209367 JAJ14035.1 GDIQ01159563 JAK92162.1 GDIQ01159565 JAK92160.1 GDIP01101618 JAM02097.1 GDIQ01074987 JAN19750.1 GDIQ01074988 JAN19749.1 GDIQ01093081 JAL58645.1 GDIQ01063197 JAN31540.1 GDIP01104580 JAL99134.1 GDIP01109781 JAL93933.1
KQ977329 KYN03511.1 GL771866 EFZ09266.1 KQ981276 KYN43542.1 ADTU01004530 KQ976444 KYM86227.1 KK107161 QOIP01000003 EZA56613.1 RLU24874.1 KQ980591 KYN15210.1 GL437344 EFN70589.1 GL888120 EGI67019.1 KQ983089 KYQ47538.1 GFDL01005773 JAV29272.1 KQ435782 KOX74771.1 KQ971361 EFA08014.1 KQ414864 KOC60003.1 GDIQ01245036 JAK06689.1 GDIQ01245035 JAK06690.1 GDIQ01064698 JAN30039.1 GDIQ01169413 JAK82312.1 GDIQ01226700 JAK25025.1 GDIQ01205298 JAK46427.1 GDIQ01135719 JAL16007.1 GDIQ01142316 JAL09410.1 GDIQ01145274 JAL06452.1 GDIQ01135718 JAL16008.1 GDIP01093029 JAM10686.1 GDIQ01135720 JAL16006.1 GDIQ01125571 GDIQ01052465 JAL26155.1 GDIQ01241140 GDIQ01125569 GDIQ01039403 JAL26157.1 GDIP01061184 JAM42531.1 GDIQ01241141 GDIQ01125570 GDIQ01066049 JAK10584.1 GDIQ01203739 JAK47986.1 GDIQ01123722 JAL28004.1 GDIP01159276 JAJ64126.1 GDIQ01235742 JAK15983.1 GDIP01152974 JAJ70428.1 GDIQ01004723 JAN90014.1 GDIP01160931 JAJ62471.1 GDIP01086430 JAM17285.1 GDIP01162420 JAJ60982.1 GDIQ01178955 JAK72770.1 GDIP01198080 JAJ25322.1 GDIP01227412 GDIP01194978 GDIP01143775 GDIP01124135 JAI95989.1 GDIP01101619 JAM02096.1 GDIQ01123721 JAL28005.1 GDIP01112436 JAL91278.1 GDIP01152071 JAJ71331.1 GDIP01216316 JAJ07086.1 GDIP01218308 JAJ05094.1 GDIP01203461 JAJ19941.1 GDIP01010066 JAM93649.1 GDIP01083434 JAM20281.1 GDIQ01155459 JAK96266.1 GDIP01218307 JAJ05095.1 GDIP01209368 JAJ14034.1 GDIQ01159564 JAK92161.1 GDIP01209367 JAJ14035.1 GDIQ01159563 JAK92162.1 GDIQ01159565 JAK92160.1 GDIP01101618 JAM02097.1 GDIQ01074987 JAN19750.1 GDIQ01074988 JAN19749.1 GDIQ01093081 JAL58645.1 GDIQ01063197 JAN31540.1 GDIP01104580 JAL99134.1 GDIP01109781 JAL93933.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JCZ3
A0A194QAK0
A0A212FAV5
A0A195CS37
E9JDB1
A0A195FTB1
+ More
A0A158NZ25 A0A195BL24 A0A026WL08 A0A151J0T2 E2A7J6 F4WFP2 A0A151WI75 A0A1Q3FP29 A0A0N0U5B8 D6WXA1 A0A0L7QN03 A0A0P5HVM8 A0A0N8ATA3 A0A0P6F6R3 A0A0P5LNU4 A0A0P5HWW3 A0A0P5J4R7 A0A0P5PEN7 A0A0P5PYN4 A0A0P5N9T1 A0A0P5NZI7 A0A0P5W1X5 A0A0P5NZF1 A0A0N8C0Y6 A0A0P5PPV8 A0A0P5Z5W1 A0A0P5I5L6 A0A0N8B815 A0A0P5PTD7 A0A0P5D8X5 A0A0P5HCD4 A0A0P5F0U4 A0A0P6IGL0 A0A0P5D4S9 A0A0P5WHD6 A0A0P5D666 A0A0N8BGW3 A0A0P5ATT2 A0A0P4YI45 A0A0P5VCJ9 A0A0P5PSY4 A0A0P5UNM9 A0A0P5DQT7 A0A0P5A513 A0A0P4ZE08 A0A0P5B8E6 A0A0P6CRP7 A0A0P5WPE5 A0A0N8BQA0 A0A0P4Z7P5 A0A1W4WNT9 A0A0P4ZUQ2 A0A0P5MSZ7 A0A0P5ARV5 A0A0N8BNT7 A0A0P5M7K6 A0A0P5VCJ7 A0A0P6DN31 A0A0N8E035 A0A0P5TDL9 A0A0P6EI51 A0A0P5V4E0 A0A0P5UQG3
A0A158NZ25 A0A195BL24 A0A026WL08 A0A151J0T2 E2A7J6 F4WFP2 A0A151WI75 A0A1Q3FP29 A0A0N0U5B8 D6WXA1 A0A0L7QN03 A0A0P5HVM8 A0A0N8ATA3 A0A0P6F6R3 A0A0P5LNU4 A0A0P5HWW3 A0A0P5J4R7 A0A0P5PEN7 A0A0P5PYN4 A0A0P5N9T1 A0A0P5NZI7 A0A0P5W1X5 A0A0P5NZF1 A0A0N8C0Y6 A0A0P5PPV8 A0A0P5Z5W1 A0A0P5I5L6 A0A0N8B815 A0A0P5PTD7 A0A0P5D8X5 A0A0P5HCD4 A0A0P5F0U4 A0A0P6IGL0 A0A0P5D4S9 A0A0P5WHD6 A0A0P5D666 A0A0N8BGW3 A0A0P5ATT2 A0A0P4YI45 A0A0P5VCJ9 A0A0P5PSY4 A0A0P5UNM9 A0A0P5DQT7 A0A0P5A513 A0A0P4ZE08 A0A0P5B8E6 A0A0P6CRP7 A0A0P5WPE5 A0A0N8BQA0 A0A0P4Z7P5 A0A1W4WNT9 A0A0P4ZUQ2 A0A0P5MSZ7 A0A0P5ARV5 A0A0N8BNT7 A0A0P5M7K6 A0A0P5VCJ7 A0A0P6DN31 A0A0N8E035 A0A0P5TDL9 A0A0P6EI51 A0A0P5V4E0 A0A0P5UQG3
Ontologies
GO
PANTHER
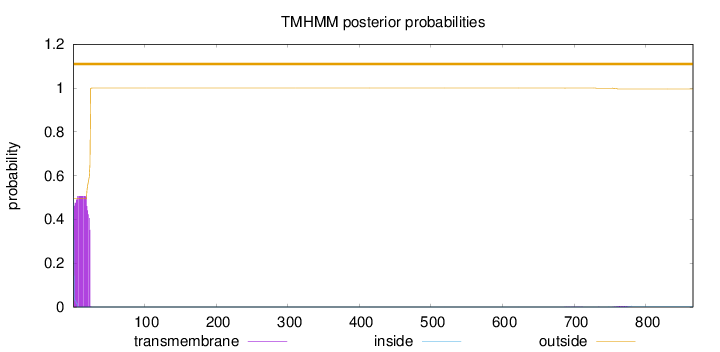
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.550819
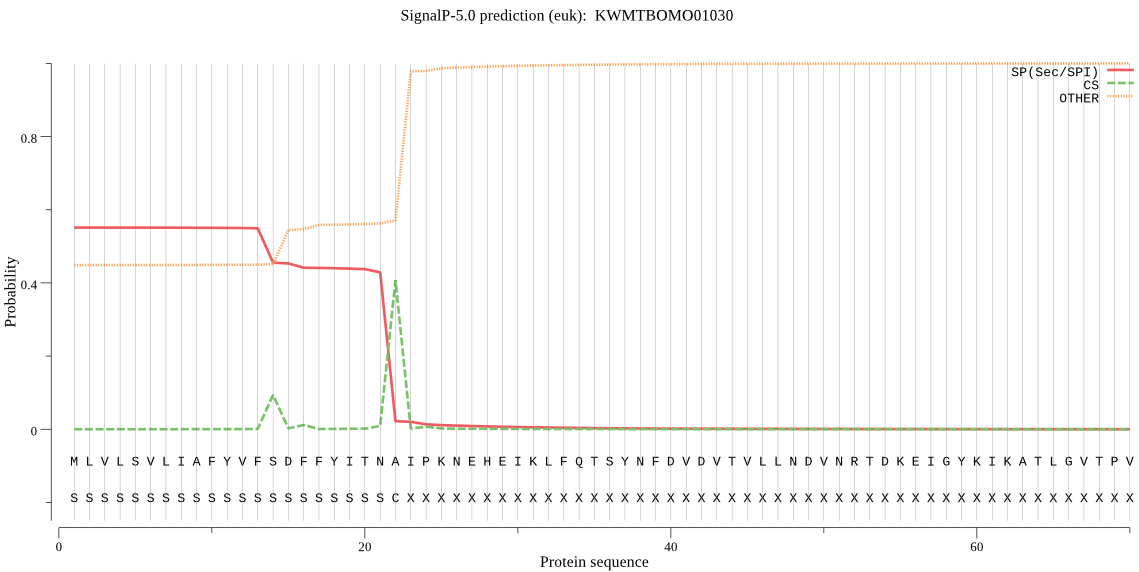
Length:
866
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.1429899999999
Exp number, first 60 AAs:
11.05883
Total prob of N-in:
0.50612
POSSIBLE N-term signal
sequence
outside
1 - 866
Population Genetic Test Statistics
Pi
196.324656
Theta
165.006556
Tajima's D
0.316426
CLR
1.334222
CSRT
0.459027048647568
Interpretation
Uncertain
