Gene
KWMTBOMO01029
Pre Gene Modal
BGIBMGA007377
Annotation
PREDICTED:_collagenase-like_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 3.155
Sequence
CDS
ATGAAACTGTTTGTACTTTGTATTGCTTTGTGCCAAGTCCTCTGGGCTTCAGGACACGTCCTGCCAGAGTTCGAGCCCATCAGCCTCAACTACCACGAGGCTGAAGGTATTCCCGCTGCAGAGCGTATCAAGCGAATTGAGGAAGCTGTGGACTTTGATGGCACCAGGATCGCAGGAGGATCCACTGCACGCTTAGGGGATCATCCTTTCCTTGCTGGATTATTGGTGCTCTTGCAAAATCGCAGGACCTCAGTGTGCGGGTCCTCTATACTTACAAATACTCGCCTGGTGACAGCAGCTCACTGCTGGTGGGATGGAACCAACCAGGGTACTGAAATTACCGCGGTCTTCGGATCCGTAACTCTCTTCTCTGGTGGCACCAGAGTCACAACCAGAACCATTGCTATGCACGGTAGCTTCAACACTCGAAACTTGAACAATGATATTGCCATCATTACTGTTCCCAGGATCAATTTCAACAACAACATTCAGCGTATAGCGATACCCAACAGCTCTCAGTCGTTCCTCTCCTTCGTCGGTAGCTGGGCTCAGTGCGCAGGCTTTGGAGTAACCAGGGATAGTGAGAACATCAACAACAATCAAGTCCTCAGACAAGTGAACCTGCAAGTGGTGAACAATATCGACTGCTCCACTGTTTATGGCTTCAATATCATAGACTCGACTCTCTGCACCAGCGGCAGGGGTAACGTTGGACCTTGCGGCGGGGACTCTGGTGGTCCCGTGTCGCTGGTGCACAGTGGCCAGCGTATCTTGATCGGTGTGGTATCATTCCACTCTGCTAACGGCTGCAACATTGGCTTCCCGGGTGGTCACGCCAGAGTTACTTCCTTCCGCTCCTGGATCAACGCACGCATTTAA
Protein
MKLFVLCIALCQVLWASGHVLPEFEPISLNYHEAEGIPAAERIKRIEEAVDFDGTRIAGGSTARLGDHPFLAGLLVLLQNRRTSVCGSSILTNTRLVTAAHCWWDGTNQGTEITAVFGSVTLFSGGTRVTTRTIAMHGSFNTRNLNNDIAIITVPRINFNNNIQRIAIPNSSQSFLSFVGSWAQCAGFGVTRDSENINNNQVLRQVNLQVVNNIDCSTVYGFNIIDSTLCTSGRGNVGPCGGDSGGPVSLVHSGQRILIGVVSFHSANGCNIGFPGGHARVTSFRSWINARI
Summary
Similarity
Belongs to the peptidase S1 family.
Belongs to the ETS family.
Belongs to the ETS family.
Uniprot
H9JCY0
A0A0N0PEU9
A0A194PUF7
A0A2W1BAG4
A0A2H1WCW9
A0A212EYA6
+ More
A0A194Q0R8 A0A0N0PA14 A0A0N0PCX7 A0A2A4JJ77 A0A0N1PGY7 H9JY12 H9JY11 Q58I80 T1WJA5 Q762I9 A0A0N1IFV7 O01953 A0A0N0PAA7 A0A0N0PAA9 Q58I79 A0A0N1PIX2 H9JY10 A0A194Q399 A0A089QFN1 A0A194Q3W8 C9W8H4 A0A0M3T9F4 H9JY13 A0A1E1WV21 A0A194R2V7 A0A0N1IAY5 S5QJ14 A0A0N1I5V5 A0A0N1PHR5 H9JY09 B2ZJV9 H9JG67 H9JJ25 Q9NH07 A0A2A4JFK9 A0A1B1LTR9 J7FCZ4 A0A1E1W2N0 A0A0M4M5D8 C9W8H9 A0PGD5 E7D003 A0A2A4JFE4 A0A1E1WB13 I6TMX9 A0A2A4JFM8 A0A1E1WL73 Q9NH10 Q1HPW8 Q9NGY4 A0A194QK37 A0A0M4LSN9 E0W9N7 A0A142I706 A0A2H1V6V4 Q25510 A0A1E1WTE4 A0A1E1WDT0 I6SJ44 I6TRU5 O18450 A0A194QCZ1 A0A2A4J8H7 A0A089RSY0 C9W8G8 H9JJ27 O18444 A0A2H1V6R8 O18438 A0A1B1LTR8 A0A2A4JGD4 A0A2A4K0D3 A0A0M5KXW7 H9JWV8 C9W8H3 A0A089RSZ3 A0A2A4JGN4 B4X988 A0A0X9H412 A0A194PHE1 B4X987 A0A0N0PBU1 A0A2A4K1Q8 A0A2A4K1C2 A0A2W1BII2 A0A2A4J8W1 B4X989 A0A1B0RHP3 A0A1B1LTR4 A0A194PHC5 E2D741 Q9N6C6 A0A2A4J6X3 A0A089QDB8
A0A194Q0R8 A0A0N0PA14 A0A0N0PCX7 A0A2A4JJ77 A0A0N1PGY7 H9JY12 H9JY11 Q58I80 T1WJA5 Q762I9 A0A0N1IFV7 O01953 A0A0N0PAA7 A0A0N0PAA9 Q58I79 A0A0N1PIX2 H9JY10 A0A194Q399 A0A089QFN1 A0A194Q3W8 C9W8H4 A0A0M3T9F4 H9JY13 A0A1E1WV21 A0A194R2V7 A0A0N1IAY5 S5QJ14 A0A0N1I5V5 A0A0N1PHR5 H9JY09 B2ZJV9 H9JG67 H9JJ25 Q9NH07 A0A2A4JFK9 A0A1B1LTR9 J7FCZ4 A0A1E1W2N0 A0A0M4M5D8 C9W8H9 A0PGD5 E7D003 A0A2A4JFE4 A0A1E1WB13 I6TMX9 A0A2A4JFM8 A0A1E1WL73 Q9NH10 Q1HPW8 Q9NGY4 A0A194QK37 A0A0M4LSN9 E0W9N7 A0A142I706 A0A2H1V6V4 Q25510 A0A1E1WTE4 A0A1E1WDT0 I6SJ44 I6TRU5 O18450 A0A194QCZ1 A0A2A4J8H7 A0A089RSY0 C9W8G8 H9JJ27 O18444 A0A2H1V6R8 O18438 A0A1B1LTR8 A0A2A4JGD4 A0A2A4K0D3 A0A0M5KXW7 H9JWV8 C9W8H3 A0A089RSZ3 A0A2A4JGN4 B4X988 A0A0X9H412 A0A194PHE1 B4X987 A0A0N0PBU1 A0A2A4K1Q8 A0A2A4K1C2 A0A2W1BII2 A0A2A4J8W1 B4X989 A0A1B0RHP3 A0A1B1LTR4 A0A194PHC5 E2D741 Q9N6C6 A0A2A4J6X3 A0A089QDB8
Pubmed
EMBL
BABH01027594
KQ459620
KPJ20544.1
KQ459591
KPI97041.1
KZ150191
+ More
PZC72392.1 ODYU01007826 SOQ50918.1 AGBW02011570 OWR46476.1 KQ459581 KPI99177.1 KQ458978 KPJ04519.1 KQ460401 KPJ14971.1 NWSH01001385 PCG71452.1 KQ461033 KPJ09997.1 BABH01041859 BABH01041860 BABH01041853 AY945210 EU338400 AAX39408.1 ABY58153.1 KF039687 AGU27161.1 AB117641 BAD13342.1 KQ460140 KPJ17495.1 BABH01042790 AB003670 BAA20136.1 KQ458725 KPJ05397.1 KPJ05400.1 AY945211 DQ310733 AAX39409.1 ABC26003.1 KPJ17496.1 BABH01041852 KQ459586 KPI97880.1 KM083780 AIR09761.1 KQ459472 KPJ00237.1 FJ205420 ACR15987.3 KR024680 ALE15222.1 BABH01041861 GDQN01000181 JAT90873.1 KQ460845 KPJ12032.1 KPJ05401.1 KC175563 AGR92346.1 KPJ05396.1 KPJ17494.1 BABH01041868 EU672968 ACD44927.1 BABH01002097 BABH01023267 AF233732 AAF71519.1 NWSH01001552 PCG70875.1 KX380993 ANS56509.1 JN252036 AFM28249.1 GDQN01009850 JAT81204.1 KR024681 ALE15223.1 FJ205425 ACR15992.2 AY618890 AAV33653.1 HM990173 ADT80821.1 NWSH01001784 PCG70143.1 GDQN01006881 JAT84173.1 JQ904138 AFM77768.1 PCG70877.1 GDQN01003329 JAT87725.1 AF233728 AAF71515.1 DQ443284 ABF51373.1 AF237417 EF531621 AAF43709.1 ABR88232.1 KQ461198 KPJ05938.1 KR024679 ALE15221.1 FJ205439 ADM35108.1 KT907053 AMR44225.1 ODYU01000971 SOQ36529.1 L34679 AAA67842.1 GDQN01000810 JAT90244.1 GDQN01005957 JAT85097.1 JQ904137 AFM77767.1 JQ904136 AFM77766.1 Y12287 CAA72966.1 KQ459185 KPJ03332.1 NWSH01002619 PCG67834.1 KM083781 AIR09762.1 FJ205413 ACR15981.1 BABH01023269 Y12280 CAA72959.1 SOQ36528.1 Y12273 CAA72952.1 KX380995 ANS56511.1 PCG70879.1 NWSH01000291 PCG77697.1 KR024677 ALE15219.1 BABH01042084 FJ205418 ACR15986.2 KM083791 AIR09772.1 SOQ36530.1 PCG70878.1 EF531624 ABR88235.1 KP690080 ALO61082.1 KQ459604 KPI92468.1 EF531623 ABR88234.1 PCG77696.1 KQ460813 KPJ12139.1 PCG77693.1 PCG77694.1 KZ150132 PZC73037.1 PCG67833.1 EF531625 ABR88236.1 KM360186 AKH49602.1 KX380990 ANS56506.1 KQ459603 KPI92811.1 GQ891130 ADK27715.1 AF233731 AF233734 AAF71518.1 AAF71716.1 PCG67835.1 KM083792 AIR09773.1
PZC72392.1 ODYU01007826 SOQ50918.1 AGBW02011570 OWR46476.1 KQ459581 KPI99177.1 KQ458978 KPJ04519.1 KQ460401 KPJ14971.1 NWSH01001385 PCG71452.1 KQ461033 KPJ09997.1 BABH01041859 BABH01041860 BABH01041853 AY945210 EU338400 AAX39408.1 ABY58153.1 KF039687 AGU27161.1 AB117641 BAD13342.1 KQ460140 KPJ17495.1 BABH01042790 AB003670 BAA20136.1 KQ458725 KPJ05397.1 KPJ05400.1 AY945211 DQ310733 AAX39409.1 ABC26003.1 KPJ17496.1 BABH01041852 KQ459586 KPI97880.1 KM083780 AIR09761.1 KQ459472 KPJ00237.1 FJ205420 ACR15987.3 KR024680 ALE15222.1 BABH01041861 GDQN01000181 JAT90873.1 KQ460845 KPJ12032.1 KPJ05401.1 KC175563 AGR92346.1 KPJ05396.1 KPJ17494.1 BABH01041868 EU672968 ACD44927.1 BABH01002097 BABH01023267 AF233732 AAF71519.1 NWSH01001552 PCG70875.1 KX380993 ANS56509.1 JN252036 AFM28249.1 GDQN01009850 JAT81204.1 KR024681 ALE15223.1 FJ205425 ACR15992.2 AY618890 AAV33653.1 HM990173 ADT80821.1 NWSH01001784 PCG70143.1 GDQN01006881 JAT84173.1 JQ904138 AFM77768.1 PCG70877.1 GDQN01003329 JAT87725.1 AF233728 AAF71515.1 DQ443284 ABF51373.1 AF237417 EF531621 AAF43709.1 ABR88232.1 KQ461198 KPJ05938.1 KR024679 ALE15221.1 FJ205439 ADM35108.1 KT907053 AMR44225.1 ODYU01000971 SOQ36529.1 L34679 AAA67842.1 GDQN01000810 JAT90244.1 GDQN01005957 JAT85097.1 JQ904137 AFM77767.1 JQ904136 AFM77766.1 Y12287 CAA72966.1 KQ459185 KPJ03332.1 NWSH01002619 PCG67834.1 KM083781 AIR09762.1 FJ205413 ACR15981.1 BABH01023269 Y12280 CAA72959.1 SOQ36528.1 Y12273 CAA72952.1 KX380995 ANS56511.1 PCG70879.1 NWSH01000291 PCG77697.1 KR024677 ALE15219.1 BABH01042084 FJ205418 ACR15986.2 KM083791 AIR09772.1 SOQ36530.1 PCG70878.1 EF531624 ABR88235.1 KP690080 ALO61082.1 KQ459604 KPI92468.1 EF531623 ABR88234.1 PCG77696.1 KQ460813 KPJ12139.1 PCG77693.1 PCG77694.1 KZ150132 PZC73037.1 PCG67833.1 EF531625 ABR88236.1 KM360186 AKH49602.1 KX380990 ANS56506.1 KQ459603 KPI92811.1 GQ891130 ADK27715.1 AF233731 AF233734 AAF71518.1 AAF71716.1 PCG67835.1 KM083792 AIR09773.1
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JCY0
A0A0N0PEU9
A0A194PUF7
A0A2W1BAG4
A0A2H1WCW9
A0A212EYA6
+ More
A0A194Q0R8 A0A0N0PA14 A0A0N0PCX7 A0A2A4JJ77 A0A0N1PGY7 H9JY12 H9JY11 Q58I80 T1WJA5 Q762I9 A0A0N1IFV7 O01953 A0A0N0PAA7 A0A0N0PAA9 Q58I79 A0A0N1PIX2 H9JY10 A0A194Q399 A0A089QFN1 A0A194Q3W8 C9W8H4 A0A0M3T9F4 H9JY13 A0A1E1WV21 A0A194R2V7 A0A0N1IAY5 S5QJ14 A0A0N1I5V5 A0A0N1PHR5 H9JY09 B2ZJV9 H9JG67 H9JJ25 Q9NH07 A0A2A4JFK9 A0A1B1LTR9 J7FCZ4 A0A1E1W2N0 A0A0M4M5D8 C9W8H9 A0PGD5 E7D003 A0A2A4JFE4 A0A1E1WB13 I6TMX9 A0A2A4JFM8 A0A1E1WL73 Q9NH10 Q1HPW8 Q9NGY4 A0A194QK37 A0A0M4LSN9 E0W9N7 A0A142I706 A0A2H1V6V4 Q25510 A0A1E1WTE4 A0A1E1WDT0 I6SJ44 I6TRU5 O18450 A0A194QCZ1 A0A2A4J8H7 A0A089RSY0 C9W8G8 H9JJ27 O18444 A0A2H1V6R8 O18438 A0A1B1LTR8 A0A2A4JGD4 A0A2A4K0D3 A0A0M5KXW7 H9JWV8 C9W8H3 A0A089RSZ3 A0A2A4JGN4 B4X988 A0A0X9H412 A0A194PHE1 B4X987 A0A0N0PBU1 A0A2A4K1Q8 A0A2A4K1C2 A0A2W1BII2 A0A2A4J8W1 B4X989 A0A1B0RHP3 A0A1B1LTR4 A0A194PHC5 E2D741 Q9N6C6 A0A2A4J6X3 A0A089QDB8
A0A194Q0R8 A0A0N0PA14 A0A0N0PCX7 A0A2A4JJ77 A0A0N1PGY7 H9JY12 H9JY11 Q58I80 T1WJA5 Q762I9 A0A0N1IFV7 O01953 A0A0N0PAA7 A0A0N0PAA9 Q58I79 A0A0N1PIX2 H9JY10 A0A194Q399 A0A089QFN1 A0A194Q3W8 C9W8H4 A0A0M3T9F4 H9JY13 A0A1E1WV21 A0A194R2V7 A0A0N1IAY5 S5QJ14 A0A0N1I5V5 A0A0N1PHR5 H9JY09 B2ZJV9 H9JG67 H9JJ25 Q9NH07 A0A2A4JFK9 A0A1B1LTR9 J7FCZ4 A0A1E1W2N0 A0A0M4M5D8 C9W8H9 A0PGD5 E7D003 A0A2A4JFE4 A0A1E1WB13 I6TMX9 A0A2A4JFM8 A0A1E1WL73 Q9NH10 Q1HPW8 Q9NGY4 A0A194QK37 A0A0M4LSN9 E0W9N7 A0A142I706 A0A2H1V6V4 Q25510 A0A1E1WTE4 A0A1E1WDT0 I6SJ44 I6TRU5 O18450 A0A194QCZ1 A0A2A4J8H7 A0A089RSY0 C9W8G8 H9JJ27 O18444 A0A2H1V6R8 O18438 A0A1B1LTR8 A0A2A4JGD4 A0A2A4K0D3 A0A0M5KXW7 H9JWV8 C9W8H3 A0A089RSZ3 A0A2A4JGN4 B4X988 A0A0X9H412 A0A194PHE1 B4X987 A0A0N0PBU1 A0A2A4K1Q8 A0A2A4K1C2 A0A2W1BII2 A0A2A4J8W1 B4X989 A0A1B0RHP3 A0A1B1LTR4 A0A194PHC5 E2D741 Q9N6C6 A0A2A4J6X3 A0A089QDB8
PDB
2HLC
E-value=1.23163e-28,
Score=313
Ontologies
GO
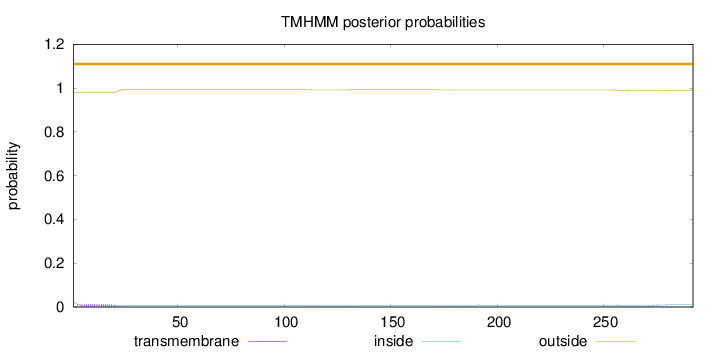
Topology
Subcellular location
Nucleus
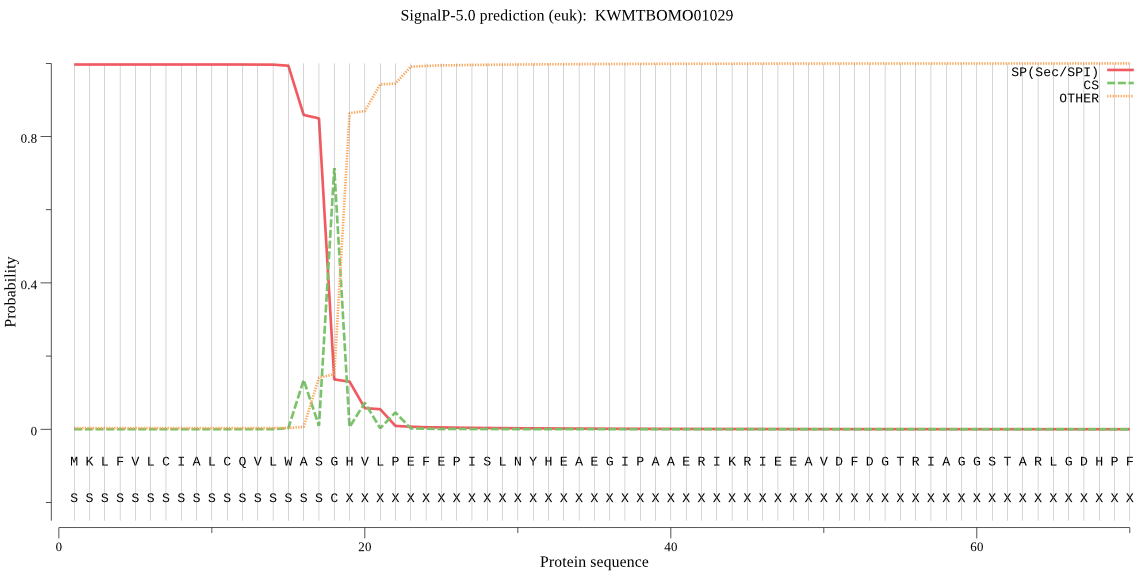
SignalP
Position: 1 - 18,
Likelihood: 0.996508
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.318560000000001
Exp number, first 60 AAs:
0.24455
Total prob of N-in:
0.02038
outside
1 - 292
Population Genetic Test Statistics
Pi
68.11483
Theta
131.3107
Tajima's D
-0.625136
CLR
6.418126
CSRT
0.212289385530723
Interpretation
Uncertain
