Gene
KWMTBOMO01010
Annotation
PREDICTED:_uncharacterized_protein_LOC101738867_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.497
Sequence
CDS
ATGTGCTTGCTCCACAGCCGCAAGTGGTCCCGCGAGCAGAAGGCGGTTGGTTCCCCAGTGTTCAACTACATGACCGCCAACACCGTGAAGACGCCGCCGGTGGGACAGCCGCCATACCAGGTGACGCTGGAGGAGCGGCTGCGGTGGGCGCAGATCGCGGAGGCCATGGCTGCGCACTACGCCCCGGAGCCGCAGAGCATGACGACGCGCCCCTCCTCCGCCATGTTCGGCGCCTACCCCACGCTACCCCCCGCGCTGCCCCCAGTGCCGCTGCCCAGGATGGGAGTCCACGGCGTGAACACGGTGACGTCACGGGCCAACACCGCCGCCTCGCACCACAACCTCTACGGGTACGGCGGGCCGCTGACGGTGGACTCGTCCGGCTCGGAGTCATCCGGGCCGCTGCCGGACCGGACCGACCTGCTGATGCCGCGCCCCAAGTCCCGCGCCAGGACCCTCCAAAGCCAGAACAACATCTACTATGACGTGGACTACGAGAACGCTCCGGAGCCGCTGTACGGCACCAAGGGCATCCCGCTGTCCACCTACAGCGTCAGCAGGGGACCGCCCTTCTACAGGCCCTGA
Protein
MCLLHSRKWSREQKAVGSPVFNYMTANTVKTPPVGQPPYQVTLEERLRWAQIAEAMAAHYAPEPQSMTTRPSSAMFGAYPTLPPALPPVPLPRMGVHGVNTVTSRANTAASHHNLYGYGGPLTVDSSGSESSGPLPDRTDLLMPRPKSRARTLQSQNNIYYDVDYENAPEPLYGTKGIPLSTYSVSRGPPFYRP
Summary
Uniprot
A0A2H1V4N4
A0A2A4JFC2
A0A2W1BH49
A0A2A4IYY1
A0A0N0PDN1
D6WQ31
+ More
A0A1B6KCZ8 A0A1B6JAF4 A0A1B6CV03 A0A1B6FG81 A0A2P8YTT1 A0A0T6BGB0 A0A023F028 A0A0A9Z2H2 A0A1B0CG44 A0A2J7PIV7 A0A1Y1LZN2 A0A2R7W1V2 A0A2H8TKY5 A0A2S2N8A2 A0A0A9Z4U5 T1I8L8 A0A1I8P3J9 A0A1I8P3L7 A0A1I8P3F8 A0A1I8P3G9 J9JM57 A0A1I8N380 A0A2S2PES4 A0A1I8P3L2 A0A2S2QSQ5 A0A1I8N378 A0A1I8P3K3 A0A336M8K0 A0A1I8N381 A0A1I8N394 A0A336L9A1 A0A336KMW4 U4UA51 A0A1A9UV57 A0A1B0FNX0 A0A1B0AAR6 A0A1A9WPB4 A0A0L0CAM1 B4J4B6 B3MI64 A0A182JX10 A0A182JGN1 A0A182LUL9 A0A182QG21 A0A182WIQ2 A0A182NHM9 B0XCT7 A0A034V5S2 E0VBN1 A0A0Q9W290 B3NA10 A0A1W4VEN8 B4KTZ7 A0A0J9TV54 A1Z6W9 A0A0R1DT23 A0A182RFM0 A0A182SSD5 B4LN72 A0A182XZ78 A0A182KRS5 A0A182I3N2 A0A0Q9W346 A0A182X6C6 A0A182U4N9 A0A182P3L5 A0A182V6B6 A0A1Y1LZP8 Q7QJQ6 B4HQS7 A0A1W4V1X5 A0A0K8WCE2 A0A084VWZ7 A0A0M3QUU1 B4MPZ4 Q17KV0 A0A182G817 A0A1S4EZ62 A0A1W4VE72 W5JDL1 A0A182FFV5 A0A034V4M7 A0A0Q5WLE8 A0A0Q9XKD8 A0A0R3NRQ5 A0A0A1WMK5 A0A1W4V1X2 B4P1M7 A0A1W4V2C8 A0A0B4KEW8
A0A1B6KCZ8 A0A1B6JAF4 A0A1B6CV03 A0A1B6FG81 A0A2P8YTT1 A0A0T6BGB0 A0A023F028 A0A0A9Z2H2 A0A1B0CG44 A0A2J7PIV7 A0A1Y1LZN2 A0A2R7W1V2 A0A2H8TKY5 A0A2S2N8A2 A0A0A9Z4U5 T1I8L8 A0A1I8P3J9 A0A1I8P3L7 A0A1I8P3F8 A0A1I8P3G9 J9JM57 A0A1I8N380 A0A2S2PES4 A0A1I8P3L2 A0A2S2QSQ5 A0A1I8N378 A0A1I8P3K3 A0A336M8K0 A0A1I8N381 A0A1I8N394 A0A336L9A1 A0A336KMW4 U4UA51 A0A1A9UV57 A0A1B0FNX0 A0A1B0AAR6 A0A1A9WPB4 A0A0L0CAM1 B4J4B6 B3MI64 A0A182JX10 A0A182JGN1 A0A182LUL9 A0A182QG21 A0A182WIQ2 A0A182NHM9 B0XCT7 A0A034V5S2 E0VBN1 A0A0Q9W290 B3NA10 A0A1W4VEN8 B4KTZ7 A0A0J9TV54 A1Z6W9 A0A0R1DT23 A0A182RFM0 A0A182SSD5 B4LN72 A0A182XZ78 A0A182KRS5 A0A182I3N2 A0A0Q9W346 A0A182X6C6 A0A182U4N9 A0A182P3L5 A0A182V6B6 A0A1Y1LZP8 Q7QJQ6 B4HQS7 A0A1W4V1X5 A0A0K8WCE2 A0A084VWZ7 A0A0M3QUU1 B4MPZ4 Q17KV0 A0A182G817 A0A1S4EZ62 A0A1W4VE72 W5JDL1 A0A182FFV5 A0A034V4M7 A0A0Q5WLE8 A0A0Q9XKD8 A0A0R3NRQ5 A0A0A1WMK5 A0A1W4V1X2 B4P1M7 A0A1W4V2C8 A0A0B4KEW8
Pubmed
28756777
26354079
18362917
19820115
29403074
25474469
+ More
25401762 28004739 25315136 23537049 26108605 17994087 25348373 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 18057021 25244985 20966253 12364791 14747013 17210077 24438588 17510324 26483478 20920257 23761445 15632085 25830018
25401762 28004739 25315136 23537049 26108605 17994087 25348373 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 18057021 25244985 20966253 12364791 14747013 17210077 24438588 17510324 26483478 20920257 23761445 15632085 25830018
EMBL
ODYU01000656
SOQ35810.1
NWSH01001596
PCG70757.1
KZ150216
PZC72136.1
+ More
NWSH01004816 PCG64676.1 KQ460296 KPJ16289.1 KQ971354 EFA06912.2 GEBQ01030648 JAT09329.1 GECU01011547 JAS96159.1 GEDC01020235 JAS17063.1 GECZ01020796 JAS48973.1 PYGN01000365 PSN47644.1 LJIG01000550 KRT86390.1 GBBI01003862 JAC14850.1 GBHO01004127 JAG39477.1 AJWK01010731 AJWK01010732 AJWK01010733 NEVH01024955 PNF16272.1 GEZM01043552 JAV79032.1 KK854237 PTY13559.1 GFXV01003002 MBW14807.1 GGMR01000553 MBY13172.1 GBHO01004125 JAG39479.1 ACPB03024601 ABLF02037374 ABLF02037375 GGMR01015286 MBY27905.1 GGMS01011551 MBY80754.1 UFQS01000481 UFQT01000481 SSX04312.1 SSX24677.1 UFQS01002141 UFQT01002141 SSX13307.1 SSX32743.1 SSX04313.1 SSX24678.1 KB632194 ERL89902.1 CCAG010009694 JRES01000678 KNC29302.1 CH916367 EDW01598.1 CH902619 EDV35909.2 AXCM01000215 AXCN02000593 DS232718 EDS45104.1 GAKP01021465 JAC37487.1 DS235033 EEB10787.1 CH940648 KRF79225.1 CH954177 EDV59706.2 CH933808 EDW08574.2 CM002911 KMY91940.1 AE013599 AAF59274.3 CM000157 KRJ98233.1 EDW60076.2 KRF79223.1 KRF79224.1 APCN01000215 KRF79222.1 GEZM01043551 JAV79033.1 AAAB01008807 EAA03940.4 CH480816 EDW46737.1 GDHF01003498 JAI48816.1 ATLV01017810 KE525192 KFB42491.1 CP012524 ALC41223.1 CH963849 EDW74183.1 CH477221 EAT47304.1 JXUM01007963 JXUM01007964 KQ560250 KXJ83566.1 ADMH02001692 ETN61413.1 GAKP01021463 JAC37489.1 KQS70969.1 KRG04179.1 CM000071 KRT01539.1 GBXI01014200 JAD00092.1 EDW89163.2 AGB93296.1
NWSH01004816 PCG64676.1 KQ460296 KPJ16289.1 KQ971354 EFA06912.2 GEBQ01030648 JAT09329.1 GECU01011547 JAS96159.1 GEDC01020235 JAS17063.1 GECZ01020796 JAS48973.1 PYGN01000365 PSN47644.1 LJIG01000550 KRT86390.1 GBBI01003862 JAC14850.1 GBHO01004127 JAG39477.1 AJWK01010731 AJWK01010732 AJWK01010733 NEVH01024955 PNF16272.1 GEZM01043552 JAV79032.1 KK854237 PTY13559.1 GFXV01003002 MBW14807.1 GGMR01000553 MBY13172.1 GBHO01004125 JAG39479.1 ACPB03024601 ABLF02037374 ABLF02037375 GGMR01015286 MBY27905.1 GGMS01011551 MBY80754.1 UFQS01000481 UFQT01000481 SSX04312.1 SSX24677.1 UFQS01002141 UFQT01002141 SSX13307.1 SSX32743.1 SSX04313.1 SSX24678.1 KB632194 ERL89902.1 CCAG010009694 JRES01000678 KNC29302.1 CH916367 EDW01598.1 CH902619 EDV35909.2 AXCM01000215 AXCN02000593 DS232718 EDS45104.1 GAKP01021465 JAC37487.1 DS235033 EEB10787.1 CH940648 KRF79225.1 CH954177 EDV59706.2 CH933808 EDW08574.2 CM002911 KMY91940.1 AE013599 AAF59274.3 CM000157 KRJ98233.1 EDW60076.2 KRF79223.1 KRF79224.1 APCN01000215 KRF79222.1 GEZM01043551 JAV79033.1 AAAB01008807 EAA03940.4 CH480816 EDW46737.1 GDHF01003498 JAI48816.1 ATLV01017810 KE525192 KFB42491.1 CP012524 ALC41223.1 CH963849 EDW74183.1 CH477221 EAT47304.1 JXUM01007963 JXUM01007964 KQ560250 KXJ83566.1 ADMH02001692 ETN61413.1 GAKP01021463 JAC37489.1 KQS70969.1 KRG04179.1 CM000071 KRT01539.1 GBXI01014200 JAD00092.1 EDW89163.2 AGB93296.1
Proteomes
UP000218220
UP000053240
UP000007266
UP000245037
UP000092461
UP000235965
+ More
UP000015103 UP000095300 UP000007819 UP000095301 UP000030742 UP000078200 UP000092444 UP000092445 UP000091820 UP000037069 UP000001070 UP000007801 UP000075881 UP000075880 UP000075883 UP000075886 UP000075920 UP000075884 UP000002320 UP000009046 UP000008792 UP000008711 UP000192221 UP000009192 UP000000803 UP000002282 UP000075900 UP000075901 UP000076408 UP000075882 UP000075840 UP000076407 UP000075902 UP000075885 UP000075903 UP000007062 UP000001292 UP000030765 UP000092553 UP000007798 UP000008820 UP000069940 UP000249989 UP000000673 UP000069272 UP000001819
UP000015103 UP000095300 UP000007819 UP000095301 UP000030742 UP000078200 UP000092444 UP000092445 UP000091820 UP000037069 UP000001070 UP000007801 UP000075881 UP000075880 UP000075883 UP000075886 UP000075920 UP000075884 UP000002320 UP000009046 UP000008792 UP000008711 UP000192221 UP000009192 UP000000803 UP000002282 UP000075900 UP000075901 UP000076408 UP000075882 UP000075840 UP000076407 UP000075902 UP000075885 UP000075903 UP000007062 UP000001292 UP000030765 UP000092553 UP000007798 UP000008820 UP000069940 UP000249989 UP000000673 UP000069272 UP000001819
PRIDE
Interpro
ProteinModelPortal
A0A2H1V4N4
A0A2A4JFC2
A0A2W1BH49
A0A2A4IYY1
A0A0N0PDN1
D6WQ31
+ More
A0A1B6KCZ8 A0A1B6JAF4 A0A1B6CV03 A0A1B6FG81 A0A2P8YTT1 A0A0T6BGB0 A0A023F028 A0A0A9Z2H2 A0A1B0CG44 A0A2J7PIV7 A0A1Y1LZN2 A0A2R7W1V2 A0A2H8TKY5 A0A2S2N8A2 A0A0A9Z4U5 T1I8L8 A0A1I8P3J9 A0A1I8P3L7 A0A1I8P3F8 A0A1I8P3G9 J9JM57 A0A1I8N380 A0A2S2PES4 A0A1I8P3L2 A0A2S2QSQ5 A0A1I8N378 A0A1I8P3K3 A0A336M8K0 A0A1I8N381 A0A1I8N394 A0A336L9A1 A0A336KMW4 U4UA51 A0A1A9UV57 A0A1B0FNX0 A0A1B0AAR6 A0A1A9WPB4 A0A0L0CAM1 B4J4B6 B3MI64 A0A182JX10 A0A182JGN1 A0A182LUL9 A0A182QG21 A0A182WIQ2 A0A182NHM9 B0XCT7 A0A034V5S2 E0VBN1 A0A0Q9W290 B3NA10 A0A1W4VEN8 B4KTZ7 A0A0J9TV54 A1Z6W9 A0A0R1DT23 A0A182RFM0 A0A182SSD5 B4LN72 A0A182XZ78 A0A182KRS5 A0A182I3N2 A0A0Q9W346 A0A182X6C6 A0A182U4N9 A0A182P3L5 A0A182V6B6 A0A1Y1LZP8 Q7QJQ6 B4HQS7 A0A1W4V1X5 A0A0K8WCE2 A0A084VWZ7 A0A0M3QUU1 B4MPZ4 Q17KV0 A0A182G817 A0A1S4EZ62 A0A1W4VE72 W5JDL1 A0A182FFV5 A0A034V4M7 A0A0Q5WLE8 A0A0Q9XKD8 A0A0R3NRQ5 A0A0A1WMK5 A0A1W4V1X2 B4P1M7 A0A1W4V2C8 A0A0B4KEW8
A0A1B6KCZ8 A0A1B6JAF4 A0A1B6CV03 A0A1B6FG81 A0A2P8YTT1 A0A0T6BGB0 A0A023F028 A0A0A9Z2H2 A0A1B0CG44 A0A2J7PIV7 A0A1Y1LZN2 A0A2R7W1V2 A0A2H8TKY5 A0A2S2N8A2 A0A0A9Z4U5 T1I8L8 A0A1I8P3J9 A0A1I8P3L7 A0A1I8P3F8 A0A1I8P3G9 J9JM57 A0A1I8N380 A0A2S2PES4 A0A1I8P3L2 A0A2S2QSQ5 A0A1I8N378 A0A1I8P3K3 A0A336M8K0 A0A1I8N381 A0A1I8N394 A0A336L9A1 A0A336KMW4 U4UA51 A0A1A9UV57 A0A1B0FNX0 A0A1B0AAR6 A0A1A9WPB4 A0A0L0CAM1 B4J4B6 B3MI64 A0A182JX10 A0A182JGN1 A0A182LUL9 A0A182QG21 A0A182WIQ2 A0A182NHM9 B0XCT7 A0A034V5S2 E0VBN1 A0A0Q9W290 B3NA10 A0A1W4VEN8 B4KTZ7 A0A0J9TV54 A1Z6W9 A0A0R1DT23 A0A182RFM0 A0A182SSD5 B4LN72 A0A182XZ78 A0A182KRS5 A0A182I3N2 A0A0Q9W346 A0A182X6C6 A0A182U4N9 A0A182P3L5 A0A182V6B6 A0A1Y1LZP8 Q7QJQ6 B4HQS7 A0A1W4V1X5 A0A0K8WCE2 A0A084VWZ7 A0A0M3QUU1 B4MPZ4 Q17KV0 A0A182G817 A0A1S4EZ62 A0A1W4VE72 W5JDL1 A0A182FFV5 A0A034V4M7 A0A0Q5WLE8 A0A0Q9XKD8 A0A0R3NRQ5 A0A0A1WMK5 A0A1W4V1X2 B4P1M7 A0A1W4V2C8 A0A0B4KEW8
Ontologies
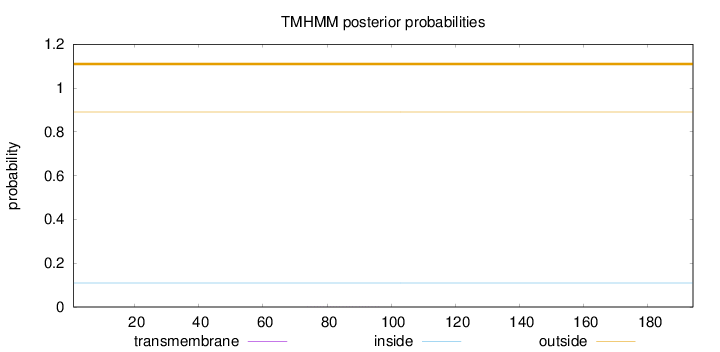
Topology
Length:
194
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00627
Exp number, first 60 AAs:
0.000620000000000001
Total prob of N-in:
0.10948
outside
1 - 194
Population Genetic Test Statistics
Pi
308.533812
Theta
165.637566
Tajima's D
2.585237
CLR
0.040947
CSRT
0.952052397380131
Interpretation
Uncertain
