Gene
KWMTBOMO01007
Pre Gene Modal
BGIBMGA001344
Annotation
cyclin_dependent_kinase_4_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.477 Mitochondrial Reliability : 1.206 Nuclear Reliability : 1.123
Sequence
CDS
ATGATGTCGCGACCGAGTACCTCACAGTCACCACCTGTTGTTGCTTCCATTAACGATGTCAGTGCATTATTTCAAAATGCACAGAAATATGAAGAACTGAGTCTAATTGGTACAGGTGCATATGGGACAGTGTATAAAGCTCGCGATCTTCATAATGGAGGGCAGATTGTAGCTATGAAGAAAGTTAAAGTTGCCCTCACAGAGGATGGGATACCACTCTCAACTGTACGAGAGATTGCCTTGTTAAGACAACTTGAGGCGTATAGACATCCCAATATAGTCAGACTGCTAGATGTCTGCCACGGAGGGCAGTGCTTGGAGCGAGACCAGCAGCTAGTACTATTCCTGGTGTTTGAACATGTGGAACAGGACCTTGACTCTTTCCTTAGTCGAGCACCGGGACCGCTTCCAGAGAGCAGGATTAGAAGCATGTCCCACGATATAATATCAGGGGTGGACTTCTTGCACTCTCATCGCATTGTGCACCGAGATCTCAAACCTCACAATCTCCTGGTGACGGTGGCTGGACGAGTCAAGCTAGCTGACTTTGGTCTCGCCAAGACCTATGATGTAGAGATGAAGCTCACCAGTGTTGTGGTGACGCTGTGGTACCGGCCCCCGGAGGTGCTGCTGGGGGCGCGCTACTGCTCGGCGCTGGACGTGTGGTCGGCGGGCTGCGTGCTGGCGCAGCTGCACACGCGCCGCCCGCTGCTGCCCGGCGCCTCCGACTCCGATCAGCTGCACCGTATCTTTAGATTAATAGGTCGGCCCCCTCGCTCGGAGTGGCCTGACAACGTATCCATAGAGCTGGACAGCTTCCCCAGCTACCCTCCGCAGGACCTGGCCAGAATCCTGCCCAGGATACATCCTCATGCATTAGATCTTATCAAGGGCATGCTGGTGTTTGACCCGTCCAAACGGCTGACGGCGATGGACTGTCTGGAGCACCCTTACTTCACGGAGGAGCCGGTCGCGTAG
Protein
MMSRPSTSQSPPVVASINDVSALFQNAQKYEELSLIGTGAYGTVYKARDLHNGGQIVAMKKVKVALTEDGIPLSTVREIALLRQLEAYRHPNIVRLLDVCHGGQCLERDQQLVLFLVFEHVEQDLDSFLSRAPGPLPESRIRSMSHDIISGVDFLHSHRIVHRDLKPHNLLVTVAGRVKLADFGLAKTYDVEMKLTSVVVTLWYRPPEVLLGARYCSALDVWSAGCVLAQLHTRRPLLPGASDSDQLHRIFRLIGRPPRSEWPDNVSIELDSFPSYPPQDLARILPRIHPHALDLIKGMLVFDPSKRLTAMDCLEHPYFTEEPVA
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
D2IGZ3
A0A2W1BE77
A0A194PTM9
A0A2H1W6R1
A0A212F2E3
A0A0N1IPK0
+ More
A0A212FE42 A0A2A4IZ57 H9IVR3 Q17L94 A0A182H2N0 B0X906 A0A1Q3FAZ0 U5EVA6 A0A0L7R6K4 A0A182FDW0 A0A084WDE3 A0A182QP23 A0A182NJH3 A0A2M4BTZ2 A0A182JK13 A0A182V4Z2 A0A182TRF8 A0A182XMG3 A0A182KQ23 Q7Q6I9 A0A182KDC4 A0A182I293 W5JMI6 A0A182S9Z4 A0A182R469 A0A182MCA2 A0A2R7WCP1 A0A182YJA6 I6LL49 A0A1W4WV22 T1KLU5 J3JXF4 A0A088AA26 A0A023F4F7 E1ZW69 A0A336K1P0 A0A224XG28 A0A291S6U9 A0A0C9R6W6 A0A1Y1L7U6 A0A0P5XSQ2 A0A026WDZ8 A0A0P5NF85 A0A3L8E4E0 A0A2S2N6M6 A0A0N0BKF9 A0A0P6H955 E9IQJ8 F4X719 A0A2H8TM23 A0A1B6L8Y6 A0A2A3EHD2 A0A151INF8 A0A3M6U1K1 J9JY47 A0A195F8R9 D6WTL2 E0VBN2 A0A151WS65 S4RCS5 A0A2B4SXB2 A0A0L0BLW6 A0A1I8MXG1 A0A1B6J566 A0A0A1WHN4 A0A1B6H2H0 A0A195DUA0 A0A3B0JGI9 A0A232EUP5 W8BJJ2 A0A0P6B8V4 A0A3B0JLS1 A0A154P6Z5 A0A0P5ST91 A0A0N8CZY7 A0A0T6BCP7 A0A087TPE2 A0A1B6CJI3 A0A1I8PNC1 A0A1D2N3C7 V3ZR16 A0A067R1E1 A0A1W4UYA1 A0A0P5RES4 A0A0K8U1N2 A0A034WGA6 A0A3Q0IND4 A0A2L2YGV4 B4MNU4 B4P5Y5 A0A1B0FMX0 A0A1A9Y2Y4 A0A1B0B069 H3DK77
A0A212FE42 A0A2A4IZ57 H9IVR3 Q17L94 A0A182H2N0 B0X906 A0A1Q3FAZ0 U5EVA6 A0A0L7R6K4 A0A182FDW0 A0A084WDE3 A0A182QP23 A0A182NJH3 A0A2M4BTZ2 A0A182JK13 A0A182V4Z2 A0A182TRF8 A0A182XMG3 A0A182KQ23 Q7Q6I9 A0A182KDC4 A0A182I293 W5JMI6 A0A182S9Z4 A0A182R469 A0A182MCA2 A0A2R7WCP1 A0A182YJA6 I6LL49 A0A1W4WV22 T1KLU5 J3JXF4 A0A088AA26 A0A023F4F7 E1ZW69 A0A336K1P0 A0A224XG28 A0A291S6U9 A0A0C9R6W6 A0A1Y1L7U6 A0A0P5XSQ2 A0A026WDZ8 A0A0P5NF85 A0A3L8E4E0 A0A2S2N6M6 A0A0N0BKF9 A0A0P6H955 E9IQJ8 F4X719 A0A2H8TM23 A0A1B6L8Y6 A0A2A3EHD2 A0A151INF8 A0A3M6U1K1 J9JY47 A0A195F8R9 D6WTL2 E0VBN2 A0A151WS65 S4RCS5 A0A2B4SXB2 A0A0L0BLW6 A0A1I8MXG1 A0A1B6J566 A0A0A1WHN4 A0A1B6H2H0 A0A195DUA0 A0A3B0JGI9 A0A232EUP5 W8BJJ2 A0A0P6B8V4 A0A3B0JLS1 A0A154P6Z5 A0A0P5ST91 A0A0N8CZY7 A0A0T6BCP7 A0A087TPE2 A0A1B6CJI3 A0A1I8PNC1 A0A1D2N3C7 V3ZR16 A0A067R1E1 A0A1W4UYA1 A0A0P5RES4 A0A0K8U1N2 A0A034WGA6 A0A3Q0IND4 A0A2L2YGV4 B4MNU4 B4P5Y5 A0A1B0FMX0 A0A1A9Y2Y4 A0A1B0B069 H3DK77
Pubmed
28756777
26354079
22118469
19121390
17510324
26483478
+ More
24438588 20966253 12364791 14747013 17210077 20920257 23761445 25244985 22516182 25474469 20798317 28992199 28004739 24508170 30249741 21282665 21719571 30382153 18362917 19820115 20566863 26108605 25315136 25830018 28648823 24495485 27289101 23254933 24845553 25348373 26561354 17994087 17550304 15496914
24438588 20966253 12364791 14747013 17210077 20920257 23761445 25244985 22516182 25474469 20798317 28992199 28004739 24508170 30249741 21282665 21719571 30382153 18362917 19820115 20566863 26108605 25315136 25830018 28648823 24495485 27289101 23254933 24845553 25348373 26561354 17994087 17550304 15496914
EMBL
GQ250575
ACT83400.1
KZ150216
PZC72135.1
KQ459593
KPI96672.1
+ More
ODYU01006684 SOQ48748.1 AGBW02010749 OWR47918.1 KQ460296 KPJ16290.1 AGBW02008983 OWR51978.1 NWSH01004335 PCG65217.1 BABH01038101 CH477217 EAT47481.1 JXUM01023842 KQ560673 KXJ81334.1 DS232513 EDS42912.1 GFDL01010301 JAV24744.1 GANO01001962 JAB57909.1 KQ414646 KOC66478.1 ATLV01022998 KE525339 KFB48237.1 AXCN02000446 GGFJ01007409 MBW56550.1 AAAB01008960 EAA10812.3 APCN01000131 ADMH02001139 ETN63964.1 AXCM01007830 KK854464 PTY16065.1 FJ966898 ADI52631.1 CAEY01000212 BT127923 AEE62885.1 GBBI01002808 JAC15904.1 GL434776 EFN74578.1 UFQS01000063 UFQT01000063 SSW98874.1 SSX19260.1 GFTR01004990 JAW11436.1 MF432983 ATL75334.1 GBYB01003695 JAG73462.1 GEZM01068144 JAV67147.1 GDIP01080091 JAM23624.1 KK107260 EZA54138.1 GDIQ01142930 JAL08796.1 QOIP01000001 RLU27343.1 GGMR01000245 MBY12864.1 KQ435700 KOX80538.1 GDIQ01022663 JAN72074.1 GL764834 EFZ17141.1 GL888828 EGI57781.1 GFXV01002907 MBW14712.1 GEBQ01027935 GEBQ01019817 JAT12042.1 JAT20160.1 KZ288249 PBC31118.1 KQ976933 KYN07010.1 RCHS01002410 RMX47575.1 ABLF02039607 KQ981744 KYN36444.1 KQ971354 EFA05832.1 DS235033 EEB10788.1 KQ982776 KYQ50749.1 LSMT01000008 PFX33819.1 JRES01001669 KNC21017.1 GECU01013399 JAS94307.1 GBXI01016262 GBXI01015985 GBXI01013775 GBXI01009918 GBXI01007336 JAC98029.1 JAC98306.1 JAD00517.1 JAD04374.1 JAD06956.1 GECZ01000904 JAS68865.1 KQ980341 KYN16431.1 OUUW01000001 SPP74470.1 NNAY01002107 OXU22064.1 GAMC01009377 JAB97178.1 GDIP01020219 JAM83496.1 SPP74469.1 KQ434822 KZC07114.1 GDIP01140953 JAL62761.1 GDIP01082351 JAM21364.1 LJIG01001840 KRT85100.1 KK116158 KFM66981.1 GEDC01023813 GEDC01017389 JAS13485.1 JAS19909.1 LJIJ01000255 ODM99786.1 KB202990 ESO86807.1 KK853098 KDR11403.1 GDIQ01101642 JAL50084.1 GDHF01031655 GDHF01021009 GDHF01007615 JAI20659.1 JAI31305.1 JAI44699.1 GAKP01005254 GAKP01005253 JAC53699.1 IAAA01025282 LAA07348.1 CH963848 EDW73783.2 CM000158 EDW91901.1 KRK00197.1 KRK00198.1 CCAG010017066 JXJN01006585 JXJN01006586
ODYU01006684 SOQ48748.1 AGBW02010749 OWR47918.1 KQ460296 KPJ16290.1 AGBW02008983 OWR51978.1 NWSH01004335 PCG65217.1 BABH01038101 CH477217 EAT47481.1 JXUM01023842 KQ560673 KXJ81334.1 DS232513 EDS42912.1 GFDL01010301 JAV24744.1 GANO01001962 JAB57909.1 KQ414646 KOC66478.1 ATLV01022998 KE525339 KFB48237.1 AXCN02000446 GGFJ01007409 MBW56550.1 AAAB01008960 EAA10812.3 APCN01000131 ADMH02001139 ETN63964.1 AXCM01007830 KK854464 PTY16065.1 FJ966898 ADI52631.1 CAEY01000212 BT127923 AEE62885.1 GBBI01002808 JAC15904.1 GL434776 EFN74578.1 UFQS01000063 UFQT01000063 SSW98874.1 SSX19260.1 GFTR01004990 JAW11436.1 MF432983 ATL75334.1 GBYB01003695 JAG73462.1 GEZM01068144 JAV67147.1 GDIP01080091 JAM23624.1 KK107260 EZA54138.1 GDIQ01142930 JAL08796.1 QOIP01000001 RLU27343.1 GGMR01000245 MBY12864.1 KQ435700 KOX80538.1 GDIQ01022663 JAN72074.1 GL764834 EFZ17141.1 GL888828 EGI57781.1 GFXV01002907 MBW14712.1 GEBQ01027935 GEBQ01019817 JAT12042.1 JAT20160.1 KZ288249 PBC31118.1 KQ976933 KYN07010.1 RCHS01002410 RMX47575.1 ABLF02039607 KQ981744 KYN36444.1 KQ971354 EFA05832.1 DS235033 EEB10788.1 KQ982776 KYQ50749.1 LSMT01000008 PFX33819.1 JRES01001669 KNC21017.1 GECU01013399 JAS94307.1 GBXI01016262 GBXI01015985 GBXI01013775 GBXI01009918 GBXI01007336 JAC98029.1 JAC98306.1 JAD00517.1 JAD04374.1 JAD06956.1 GECZ01000904 JAS68865.1 KQ980341 KYN16431.1 OUUW01000001 SPP74470.1 NNAY01002107 OXU22064.1 GAMC01009377 JAB97178.1 GDIP01020219 JAM83496.1 SPP74469.1 KQ434822 KZC07114.1 GDIP01140953 JAL62761.1 GDIP01082351 JAM21364.1 LJIG01001840 KRT85100.1 KK116158 KFM66981.1 GEDC01023813 GEDC01017389 JAS13485.1 JAS19909.1 LJIJ01000255 ODM99786.1 KB202990 ESO86807.1 KK853098 KDR11403.1 GDIQ01101642 JAL50084.1 GDHF01031655 GDHF01021009 GDHF01007615 JAI20659.1 JAI31305.1 JAI44699.1 GAKP01005254 GAKP01005253 JAC53699.1 IAAA01025282 LAA07348.1 CH963848 EDW73783.2 CM000158 EDW91901.1 KRK00197.1 KRK00198.1 CCAG010017066 JXJN01006585 JXJN01006586
Proteomes
UP000053268
UP000007151
UP000053240
UP000218220
UP000005204
UP000008820
+ More
UP000069940 UP000249989 UP000002320 UP000053825 UP000069272 UP000030765 UP000075886 UP000075884 UP000075880 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075881 UP000075840 UP000000673 UP000075901 UP000075900 UP000075883 UP000076408 UP000192223 UP000015104 UP000005203 UP000000311 UP000053097 UP000279307 UP000053105 UP000007755 UP000242457 UP000078542 UP000275408 UP000007819 UP000078541 UP000007266 UP000009046 UP000075809 UP000245300 UP000225706 UP000037069 UP000095301 UP000078492 UP000268350 UP000215335 UP000076502 UP000054359 UP000095300 UP000094527 UP000030746 UP000027135 UP000192221 UP000079169 UP000007798 UP000002282 UP000092444 UP000092443 UP000092460 UP000007303
UP000069940 UP000249989 UP000002320 UP000053825 UP000069272 UP000030765 UP000075886 UP000075884 UP000075880 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075881 UP000075840 UP000000673 UP000075901 UP000075900 UP000075883 UP000076408 UP000192223 UP000015104 UP000005203 UP000000311 UP000053097 UP000279307 UP000053105 UP000007755 UP000242457 UP000078542 UP000275408 UP000007819 UP000078541 UP000007266 UP000009046 UP000075809 UP000245300 UP000225706 UP000037069 UP000095301 UP000078492 UP000268350 UP000215335 UP000076502 UP000054359 UP000095300 UP000094527 UP000030746 UP000027135 UP000192221 UP000079169 UP000007798 UP000002282 UP000092444 UP000092443 UP000092460 UP000007303
Interpro
Gene 3D
ProteinModelPortal
D2IGZ3
A0A2W1BE77
A0A194PTM9
A0A2H1W6R1
A0A212F2E3
A0A0N1IPK0
+ More
A0A212FE42 A0A2A4IZ57 H9IVR3 Q17L94 A0A182H2N0 B0X906 A0A1Q3FAZ0 U5EVA6 A0A0L7R6K4 A0A182FDW0 A0A084WDE3 A0A182QP23 A0A182NJH3 A0A2M4BTZ2 A0A182JK13 A0A182V4Z2 A0A182TRF8 A0A182XMG3 A0A182KQ23 Q7Q6I9 A0A182KDC4 A0A182I293 W5JMI6 A0A182S9Z4 A0A182R469 A0A182MCA2 A0A2R7WCP1 A0A182YJA6 I6LL49 A0A1W4WV22 T1KLU5 J3JXF4 A0A088AA26 A0A023F4F7 E1ZW69 A0A336K1P0 A0A224XG28 A0A291S6U9 A0A0C9R6W6 A0A1Y1L7U6 A0A0P5XSQ2 A0A026WDZ8 A0A0P5NF85 A0A3L8E4E0 A0A2S2N6M6 A0A0N0BKF9 A0A0P6H955 E9IQJ8 F4X719 A0A2H8TM23 A0A1B6L8Y6 A0A2A3EHD2 A0A151INF8 A0A3M6U1K1 J9JY47 A0A195F8R9 D6WTL2 E0VBN2 A0A151WS65 S4RCS5 A0A2B4SXB2 A0A0L0BLW6 A0A1I8MXG1 A0A1B6J566 A0A0A1WHN4 A0A1B6H2H0 A0A195DUA0 A0A3B0JGI9 A0A232EUP5 W8BJJ2 A0A0P6B8V4 A0A3B0JLS1 A0A154P6Z5 A0A0P5ST91 A0A0N8CZY7 A0A0T6BCP7 A0A087TPE2 A0A1B6CJI3 A0A1I8PNC1 A0A1D2N3C7 V3ZR16 A0A067R1E1 A0A1W4UYA1 A0A0P5RES4 A0A0K8U1N2 A0A034WGA6 A0A3Q0IND4 A0A2L2YGV4 B4MNU4 B4P5Y5 A0A1B0FMX0 A0A1A9Y2Y4 A0A1B0B069 H3DK77
A0A212FE42 A0A2A4IZ57 H9IVR3 Q17L94 A0A182H2N0 B0X906 A0A1Q3FAZ0 U5EVA6 A0A0L7R6K4 A0A182FDW0 A0A084WDE3 A0A182QP23 A0A182NJH3 A0A2M4BTZ2 A0A182JK13 A0A182V4Z2 A0A182TRF8 A0A182XMG3 A0A182KQ23 Q7Q6I9 A0A182KDC4 A0A182I293 W5JMI6 A0A182S9Z4 A0A182R469 A0A182MCA2 A0A2R7WCP1 A0A182YJA6 I6LL49 A0A1W4WV22 T1KLU5 J3JXF4 A0A088AA26 A0A023F4F7 E1ZW69 A0A336K1P0 A0A224XG28 A0A291S6U9 A0A0C9R6W6 A0A1Y1L7U6 A0A0P5XSQ2 A0A026WDZ8 A0A0P5NF85 A0A3L8E4E0 A0A2S2N6M6 A0A0N0BKF9 A0A0P6H955 E9IQJ8 F4X719 A0A2H8TM23 A0A1B6L8Y6 A0A2A3EHD2 A0A151INF8 A0A3M6U1K1 J9JY47 A0A195F8R9 D6WTL2 E0VBN2 A0A151WS65 S4RCS5 A0A2B4SXB2 A0A0L0BLW6 A0A1I8MXG1 A0A1B6J566 A0A0A1WHN4 A0A1B6H2H0 A0A195DUA0 A0A3B0JGI9 A0A232EUP5 W8BJJ2 A0A0P6B8V4 A0A3B0JLS1 A0A154P6Z5 A0A0P5ST91 A0A0N8CZY7 A0A0T6BCP7 A0A087TPE2 A0A1B6CJI3 A0A1I8PNC1 A0A1D2N3C7 V3ZR16 A0A067R1E1 A0A1W4UYA1 A0A0P5RES4 A0A0K8U1N2 A0A034WGA6 A0A3Q0IND4 A0A2L2YGV4 B4MNU4 B4P5Y5 A0A1B0FMX0 A0A1A9Y2Y4 A0A1B0B069 H3DK77
PDB
2F2C
E-value=5.27356e-85,
Score=800
Ontologies
PATHWAY
GO
Topology
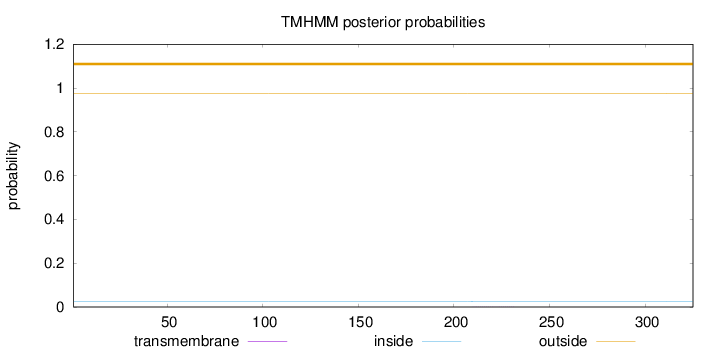
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00589
Exp number, first 60 AAs:
0.00067
Total prob of N-in:
0.02604
outside
1 - 325
Population Genetic Test Statistics
Pi
285.9033
Theta
146.199396
Tajima's D
2.819769
CLR
0.092872
CSRT
0.969551522423879
Interpretation
Uncertain
