Gene
KWMTBOMO00999
Annotation
PREDICTED:_LYR_motif-containing_protein_4_[Amyelois_transitella]
Full name
LYR motif-containing protein 4
+ More
LYR motif-containing protein 4B
Protein bcn92
LYR motif-containing protein 4B
Protein bcn92
Location in the cell
Mitochondrial Reliability : 2.142
Sequence
CDS
ATGTCTGGCGCGATACCTAAAATACAAATATTATCTCTCTATAAATTGCTTCTCAGAGAATCCCAGAAGTTCCCTAACTATAATTTCAGAGCATACGCACTAAGAAGAATTCGAGATGCTTTTAAAGATAATAAAAATGTGTCTGATGTTAAGATTGTGAAGAAAGAGTTTGAATTTGGAAAAGAAAATTTAAATGTTATCAGGAGACAGGCAGCCATTGGAAATATGTATAAAACAGAAAAACTTGTTATCGAAAATCTGCGGTAG
Protein
MSGAIPKIQILSLYKLLLRESQKFPNYNFRAYALRRIRDAFKDNKNVSDVKIVKKEFEFGKENLNVIRRQAAIGNMYKTEKLVIENLR
Summary
Description
Required for nuclear and mitochondrial iron-sulfur protein biosynthesis.
Subunit
Interacts with FXN. Interaction is increased by nickel. Interaction is inhibited by calcium, magnesium, manganese, copper, cobalt, zinc, and iron. Forms a complex with the cytosolic/nuclear form of NFS1. The complex increased the stability of NFS1.
Miscellaneous
Reduction of LYRM4 levels by siRNA increases the total iron content, and reduces cytosolic and mitochondrial aconitase activities and NFS1 protein levels.
Similarity
Belongs to the complex I LYR family.
Keywords
Mitochondrion
Nucleus
Complete proteome
Reference proteome
Coiled coil
3D-structure
Direct protein sequencing
Disease mutation
Polymorphism
Primary mitochondrial disease
Feature
chain LYR motif-containing protein 4
sequence variant In dbSNP:rs2224391.
sequence variant In dbSNP:rs2224391.
Uniprot
S4NPZ7
A0A2A4J4W8
A0A1E1WJM5
A0A2H1V2F1
A0A212EPI9
A0A194R4S4
+ More
A0A194PSP1 A0A023EBS4 A0A1B0D317 Q178E3 C1BY44 W5N120 A0A1L8FXV4 Q6DCS1 A0A1Q3FCT5 B0W311 A0A182P6Q7 A0A3B4AAZ9 U5ESJ4 A0A1L8DAM0 U3KFD8 A0A0K8TS46 A0A1S3NCR0 B5XD90 A0A1W5B7P0 A0A139WG63 E3TD00 A0A023EE00 A0A3P8QEA8 A0A3P9CXS0 A0A1D2MU10 B5FZA8 E9FZ49 A0A182ILR3 A0A3P9M4E5 R4GGF7 A0A218UX08 A0A1U7S454 A0A151NFI0 A0A2M3ZCC2 A0A3B3IJ86 A0A2I4CYP2 A0A2M4C4U6 A0A182VC15 E3TDP8 A0A2M3ZCD8 A0A1V4JWI7 A0A3B3CWU1 A0A3P8RWE4 A0A3B3SED9 A0NDE4 A0A2R8Q8D7 A0A3B4UVV0 A0A2K5JM35 A0A1I8MBU7 A0A287AUJ0 Q9HD34 M4AUT3 A0A340Y1T5 P82116 A0A067RAW7 A0A0S7HTE1 B3MSG3 A0A2M4CZC2 A0A2U4C5N2 T1DHP7 A0A2K6DQ88 A0A2K6MNX1 A0A2K5TXT0 A0A0D9R4P3 A0A2I3MAL7 A0A2K5MDQ8 H9FMX8 A0A182RYR1 A0A2Y9SKG8 A0A0E9X8C4 L8ITK0 Q0VCG0 A0A2I2Z0B2 A0A2R9A3H3 A0A2I3SE95 A0A2R7WVE7 N6TU46 A0A1A7YW81 A0A2R8MCW8 A0A384C8D0 A0A2I3H651 A0A2K6UTV4 A0A0L0C3T1 K9IGF1 A0A2Y9PAL3 A0A2K5CXV1 A0A3B3XFT7 A0A0P5XQH2 A0A286XEV2 A0A3B0JY32 A0A2Y9KHY1 M3XN76 A0A1W4Z0F0 W5JBU6
A0A194PSP1 A0A023EBS4 A0A1B0D317 Q178E3 C1BY44 W5N120 A0A1L8FXV4 Q6DCS1 A0A1Q3FCT5 B0W311 A0A182P6Q7 A0A3B4AAZ9 U5ESJ4 A0A1L8DAM0 U3KFD8 A0A0K8TS46 A0A1S3NCR0 B5XD90 A0A1W5B7P0 A0A139WG63 E3TD00 A0A023EE00 A0A3P8QEA8 A0A3P9CXS0 A0A1D2MU10 B5FZA8 E9FZ49 A0A182ILR3 A0A3P9M4E5 R4GGF7 A0A218UX08 A0A1U7S454 A0A151NFI0 A0A2M3ZCC2 A0A3B3IJ86 A0A2I4CYP2 A0A2M4C4U6 A0A182VC15 E3TDP8 A0A2M3ZCD8 A0A1V4JWI7 A0A3B3CWU1 A0A3P8RWE4 A0A3B3SED9 A0NDE4 A0A2R8Q8D7 A0A3B4UVV0 A0A2K5JM35 A0A1I8MBU7 A0A287AUJ0 Q9HD34 M4AUT3 A0A340Y1T5 P82116 A0A067RAW7 A0A0S7HTE1 B3MSG3 A0A2M4CZC2 A0A2U4C5N2 T1DHP7 A0A2K6DQ88 A0A2K6MNX1 A0A2K5TXT0 A0A0D9R4P3 A0A2I3MAL7 A0A2K5MDQ8 H9FMX8 A0A182RYR1 A0A2Y9SKG8 A0A0E9X8C4 L8ITK0 Q0VCG0 A0A2I2Z0B2 A0A2R9A3H3 A0A2I3SE95 A0A2R7WVE7 N6TU46 A0A1A7YW81 A0A2R8MCW8 A0A384C8D0 A0A2I3H651 A0A2K6UTV4 A0A0L0C3T1 K9IGF1 A0A2Y9PAL3 A0A2K5CXV1 A0A3B3XFT7 A0A0P5XQH2 A0A286XEV2 A0A3B0JY32 A0A2Y9KHY1 M3XN76 A0A1W4Z0F0 W5JBU6
Pubmed
23622113
22118469
26354079
24945155
26483478
17510324
+ More
20433749 25069045 27762356 25463417 26369729 18362917 19820115 20634964 25186727 27289101 17018643 21292972 17554307 22293439 29451363 29240929 12364791 14747013 17210077 23594743 25315136 11342225 14702039 14574404 15489334 17331979 18650437 19454487 21269460 25944712 23814038 23542700 7705632 24845553 17994087 25319552 25613341 22751099 22398555 22722832 16136131 23537049 24813606 26108605 21993624 20920257 23761445
20433749 25069045 27762356 25463417 26369729 18362917 19820115 20634964 25186727 27289101 17018643 21292972 17554307 22293439 29451363 29240929 12364791 14747013 17210077 23594743 25315136 11342225 14702039 14574404 15489334 17331979 18650437 19454487 21269460 25944712 23814038 23542700 7705632 24845553 17994087 25319552 25613341 22751099 22398555 22722832 16136131 23537049 24813606 26108605 21993624 20920257 23761445
EMBL
GAIX01013386
JAA79174.1
NWSH01003363
PCG66470.1
GDQN01003947
JAT87107.1
+ More
ODYU01000154 ODYU01010951 SOQ34444.1 SOQ56381.1 AGBW02013452 OWR43403.1 KQ460890 KPJ10861.1 KQ459593 KPI96451.1 JXUM01109088 GAPW01006666 GAPW01006665 KQ565289 JAC06933.1 KXJ71126.1 AJVK01023187 CH477365 EAT42558.1 EJY57609.1 EJY57610.1 BT079523 ACO13947.1 AHAT01031817 AHAT01031818 CM004476 OCT76404.1 BC077926 GFDL01009651 JAV25394.1 DS231830 EDS30719.1 GANO01003219 JAB56652.1 GFDF01010576 JAV03508.1 AGTO01000234 GDAI01000853 JAI16750.1 BT049009 KQ971351 KYB26845.1 GU588230 ADO28186.1 GAPW01006754 JAC06844.1 LJIJ01000555 ODM96294.1 DQ214409 GL732527 EFX87649.1 MUZQ01000101 OWK58364.1 AKHW03003201 KYO35510.1 GGFM01005456 MBW26207.1 GGFJ01011212 MBW60353.1 GU588478 ADO28434.1 GGFM01005418 MBW26169.1 LSYS01005643 OPJ76586.1 AAAB01008880 EAU76964.2 BX784022 CABZ01061196 CABZ01074958 CU104764 FQ790217 AEMK02000051 AF285118 AF170070 AK291158 AL035653 AL121978 AL162381 BC009552 Z46522 KK852816 KDR15786.1 GBYX01437054 GBYX01437053 GBYX01437052 JAO44289.1 CH902622 EDV34718.1 GGFL01006030 MBW70208.1 GAMD01002236 JAA99354.1 AQIA01052906 AQIA01052907 AQIA01052908 AQIB01159265 AQIB01159266 AHZZ02024070 AHZZ02024071 JU332232 JU473752 JV044062 AFE75987.1 AFH30556.1 AFI34133.1 GBXM01009861 JAH98716.1 JH880690 ELR59686.1 BC120188 CABD030043064 CABD030043065 CABD030043066 CABD030043067 CABD030043068 AJFE02018850 AJFE02018851 AJFE02018852 AACZ04065956 NBAG03000244 PNI62945.1 KK855635 PTY23371.1 APGK01052243 KB741211 KB632263 ENN72790.1 ERL90924.1 HADX01011973 SBP34205.1 ADFV01069846 ADFV01069847 ADFV01069848 ADFV01069849 ADFV01069850 ADFV01069851 JRES01000940 KNC26998.1 GABZ01008683 JAA44842.1 GDIP01068723 JAM34992.1 AAKN02036990 AAKN02036991 OUUW01000003 SPP77631.1 AEYP01034674 AEYP01034675 AEYP01034676 AEYP01034677 AEYP01034678 AEYP01034679 ADMH02001952 ETN60340.1
ODYU01000154 ODYU01010951 SOQ34444.1 SOQ56381.1 AGBW02013452 OWR43403.1 KQ460890 KPJ10861.1 KQ459593 KPI96451.1 JXUM01109088 GAPW01006666 GAPW01006665 KQ565289 JAC06933.1 KXJ71126.1 AJVK01023187 CH477365 EAT42558.1 EJY57609.1 EJY57610.1 BT079523 ACO13947.1 AHAT01031817 AHAT01031818 CM004476 OCT76404.1 BC077926 GFDL01009651 JAV25394.1 DS231830 EDS30719.1 GANO01003219 JAB56652.1 GFDF01010576 JAV03508.1 AGTO01000234 GDAI01000853 JAI16750.1 BT049009 KQ971351 KYB26845.1 GU588230 ADO28186.1 GAPW01006754 JAC06844.1 LJIJ01000555 ODM96294.1 DQ214409 GL732527 EFX87649.1 MUZQ01000101 OWK58364.1 AKHW03003201 KYO35510.1 GGFM01005456 MBW26207.1 GGFJ01011212 MBW60353.1 GU588478 ADO28434.1 GGFM01005418 MBW26169.1 LSYS01005643 OPJ76586.1 AAAB01008880 EAU76964.2 BX784022 CABZ01061196 CABZ01074958 CU104764 FQ790217 AEMK02000051 AF285118 AF170070 AK291158 AL035653 AL121978 AL162381 BC009552 Z46522 KK852816 KDR15786.1 GBYX01437054 GBYX01437053 GBYX01437052 JAO44289.1 CH902622 EDV34718.1 GGFL01006030 MBW70208.1 GAMD01002236 JAA99354.1 AQIA01052906 AQIA01052907 AQIA01052908 AQIB01159265 AQIB01159266 AHZZ02024070 AHZZ02024071 JU332232 JU473752 JV044062 AFE75987.1 AFH30556.1 AFI34133.1 GBXM01009861 JAH98716.1 JH880690 ELR59686.1 BC120188 CABD030043064 CABD030043065 CABD030043066 CABD030043067 CABD030043068 AJFE02018850 AJFE02018851 AJFE02018852 AACZ04065956 NBAG03000244 PNI62945.1 KK855635 PTY23371.1 APGK01052243 KB741211 KB632263 ENN72790.1 ERL90924.1 HADX01011973 SBP34205.1 ADFV01069846 ADFV01069847 ADFV01069848 ADFV01069849 ADFV01069850 ADFV01069851 JRES01000940 KNC26998.1 GABZ01008683 JAA44842.1 GDIP01068723 JAM34992.1 AAKN02036990 AAKN02036991 OUUW01000003 SPP77631.1 AEYP01034674 AEYP01034675 AEYP01034676 AEYP01034677 AEYP01034678 AEYP01034679 ADMH02001952 ETN60340.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000069940
UP000249989
+ More
UP000092462 UP000008820 UP000265140 UP000018468 UP000186698 UP000002320 UP000075885 UP000261520 UP000016665 UP000087266 UP000192224 UP000007266 UP000265100 UP000265160 UP000094527 UP000007754 UP000000305 UP000075880 UP000265180 UP000265200 UP000197619 UP000189705 UP000050525 UP000001038 UP000192220 UP000075903 UP000221080 UP000190648 UP000261560 UP000265080 UP000261540 UP000007062 UP000000437 UP000261420 UP000233080 UP000095301 UP000008227 UP000005640 UP000002852 UP000265300 UP000027135 UP000007801 UP000245320 UP000233120 UP000233180 UP000233100 UP000029965 UP000028761 UP000233060 UP000075900 UP000248484 UP000009136 UP000001519 UP000240080 UP000002277 UP000019118 UP000030742 UP000008225 UP000261680 UP000291021 UP000001073 UP000233220 UP000037069 UP000248483 UP000233020 UP000261480 UP000005447 UP000268350 UP000248482 UP000000715 UP000000673
UP000092462 UP000008820 UP000265140 UP000018468 UP000186698 UP000002320 UP000075885 UP000261520 UP000016665 UP000087266 UP000192224 UP000007266 UP000265100 UP000265160 UP000094527 UP000007754 UP000000305 UP000075880 UP000265180 UP000265200 UP000197619 UP000189705 UP000050525 UP000001038 UP000192220 UP000075903 UP000221080 UP000190648 UP000261560 UP000265080 UP000261540 UP000007062 UP000000437 UP000261420 UP000233080 UP000095301 UP000008227 UP000005640 UP000002852 UP000265300 UP000027135 UP000007801 UP000245320 UP000233120 UP000233180 UP000233100 UP000029965 UP000028761 UP000233060 UP000075900 UP000248484 UP000009136 UP000001519 UP000240080 UP000002277 UP000019118 UP000030742 UP000008225 UP000261680 UP000291021 UP000001073 UP000233220 UP000037069 UP000248483 UP000233020 UP000261480 UP000005447 UP000268350 UP000248482 UP000000715 UP000000673
Pfam
PF05347 Complex1_LYR
Interpro
IPR008011
Complex1_LYR
ProteinModelPortal
S4NPZ7
A0A2A4J4W8
A0A1E1WJM5
A0A2H1V2F1
A0A212EPI9
A0A194R4S4
+ More
A0A194PSP1 A0A023EBS4 A0A1B0D317 Q178E3 C1BY44 W5N120 A0A1L8FXV4 Q6DCS1 A0A1Q3FCT5 B0W311 A0A182P6Q7 A0A3B4AAZ9 U5ESJ4 A0A1L8DAM0 U3KFD8 A0A0K8TS46 A0A1S3NCR0 B5XD90 A0A1W5B7P0 A0A139WG63 E3TD00 A0A023EE00 A0A3P8QEA8 A0A3P9CXS0 A0A1D2MU10 B5FZA8 E9FZ49 A0A182ILR3 A0A3P9M4E5 R4GGF7 A0A218UX08 A0A1U7S454 A0A151NFI0 A0A2M3ZCC2 A0A3B3IJ86 A0A2I4CYP2 A0A2M4C4U6 A0A182VC15 E3TDP8 A0A2M3ZCD8 A0A1V4JWI7 A0A3B3CWU1 A0A3P8RWE4 A0A3B3SED9 A0NDE4 A0A2R8Q8D7 A0A3B4UVV0 A0A2K5JM35 A0A1I8MBU7 A0A287AUJ0 Q9HD34 M4AUT3 A0A340Y1T5 P82116 A0A067RAW7 A0A0S7HTE1 B3MSG3 A0A2M4CZC2 A0A2U4C5N2 T1DHP7 A0A2K6DQ88 A0A2K6MNX1 A0A2K5TXT0 A0A0D9R4P3 A0A2I3MAL7 A0A2K5MDQ8 H9FMX8 A0A182RYR1 A0A2Y9SKG8 A0A0E9X8C4 L8ITK0 Q0VCG0 A0A2I2Z0B2 A0A2R9A3H3 A0A2I3SE95 A0A2R7WVE7 N6TU46 A0A1A7YW81 A0A2R8MCW8 A0A384C8D0 A0A2I3H651 A0A2K6UTV4 A0A0L0C3T1 K9IGF1 A0A2Y9PAL3 A0A2K5CXV1 A0A3B3XFT7 A0A0P5XQH2 A0A286XEV2 A0A3B0JY32 A0A2Y9KHY1 M3XN76 A0A1W4Z0F0 W5JBU6
A0A194PSP1 A0A023EBS4 A0A1B0D317 Q178E3 C1BY44 W5N120 A0A1L8FXV4 Q6DCS1 A0A1Q3FCT5 B0W311 A0A182P6Q7 A0A3B4AAZ9 U5ESJ4 A0A1L8DAM0 U3KFD8 A0A0K8TS46 A0A1S3NCR0 B5XD90 A0A1W5B7P0 A0A139WG63 E3TD00 A0A023EE00 A0A3P8QEA8 A0A3P9CXS0 A0A1D2MU10 B5FZA8 E9FZ49 A0A182ILR3 A0A3P9M4E5 R4GGF7 A0A218UX08 A0A1U7S454 A0A151NFI0 A0A2M3ZCC2 A0A3B3IJ86 A0A2I4CYP2 A0A2M4C4U6 A0A182VC15 E3TDP8 A0A2M3ZCD8 A0A1V4JWI7 A0A3B3CWU1 A0A3P8RWE4 A0A3B3SED9 A0NDE4 A0A2R8Q8D7 A0A3B4UVV0 A0A2K5JM35 A0A1I8MBU7 A0A287AUJ0 Q9HD34 M4AUT3 A0A340Y1T5 P82116 A0A067RAW7 A0A0S7HTE1 B3MSG3 A0A2M4CZC2 A0A2U4C5N2 T1DHP7 A0A2K6DQ88 A0A2K6MNX1 A0A2K5TXT0 A0A0D9R4P3 A0A2I3MAL7 A0A2K5MDQ8 H9FMX8 A0A182RYR1 A0A2Y9SKG8 A0A0E9X8C4 L8ITK0 Q0VCG0 A0A2I2Z0B2 A0A2R9A3H3 A0A2I3SE95 A0A2R7WVE7 N6TU46 A0A1A7YW81 A0A2R8MCW8 A0A384C8D0 A0A2I3H651 A0A2K6UTV4 A0A0L0C3T1 K9IGF1 A0A2Y9PAL3 A0A2K5CXV1 A0A3B3XFT7 A0A0P5XQH2 A0A286XEV2 A0A3B0JY32 A0A2Y9KHY1 M3XN76 A0A1W4Z0F0 W5JBU6
PDB
6NZU
E-value=8.28291e-16,
Score=197
Ontologies
Topology
Subcellular location
Mitochondrion
Nucleus
Nucleus
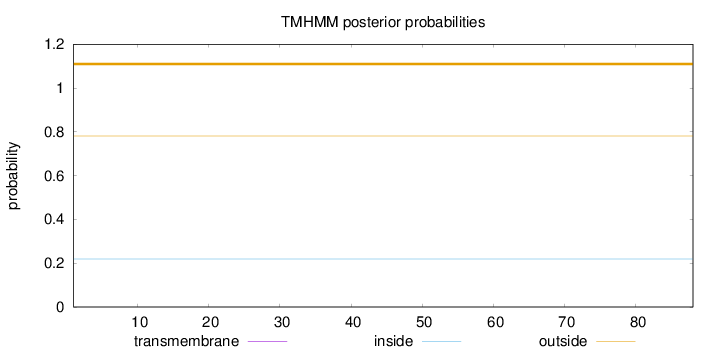
Length:
88
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00054
Exp number, first 60 AAs:
0.00054
Total prob of N-in:
0.21900
outside
1 - 88
Population Genetic Test Statistics
Pi
430.105169
Theta
210.902502
Tajima's D
3.408876
CLR
0.067463
CSRT
0.993000349982501
Interpretation
Uncertain
