Pre Gene Modal
BGIBMGA001348
Annotation
PREDICTED:_F-box/LRR-repeat_protein_20_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.543
Sequence
CDS
ATGACGCATTCTGGGAGGAGTAAATTCGAGATAACTCGTTTTACGAACGAAGATGAGAACCACATCAACAGGAAACTGCCCAAGGAGCTGCTGCTGAGGATACTGTCGTACTTAGACGTGGTGTCGCTGTGCAGATGTGCGCAGGTCTCCAAGCTGTGGAACATTCTGGCGCTGGACGGCTCCAACTGGCAACGAATAGATTTGTTCGAGTTTCAGAGGGACGTAGAGGGTCCGGTCATAGAGAACATATCGCAGCGGTGCGGCGGGTTCCTCCGGCAGCTGTCCCTCAGGGGCTGCGAGAGCATCGGCGACGGCTCCATGAAGACGCTGGCGCAGTCGTGTCCGAACATTGAAGATCTGAACCTGAACAAATGCAAGAAGATAACGGACCAGACGTGCCAGGCGCTGGGCAGGAGGTGCAGCAAACTGCAGAAAATAAATTTGGACTCGTGCCCCAGCGTGTCGGACATCTCTCTGAAGGCGCTGGCCGACGGCTGCCCTTTGCTGACACACGTGAACGTGTCGTGGTGCCAGTCCATAACGGAGAGCGGCGTGGAGGCGCTGGCCCGGGGCTGCCCCAAGCTCAAGAGCTTCATTTGTAAAGGATGCAAGAACGTCAACGACCGCGCTGTCATCAGCCTCGCCACGCGCTGCCCCGACCTCGAGGTGCTGGACATACAGGGCTGCGAGACGGTGTCGGACGAGGCGGTGTCGCGGCTGGGCGGCGCGCTGCGGCGGCTGTGCGCGTCGGGCTGCGCGCGGCTCACGGACGCGTCGCTGGCCGCGCTGGCCGCCGCCGCGCCGCGCCTCGCCACGCTCGAGCTGGCGCAGTGCACGCAGCTCACCGACGCAGGCTTCCAGGCGCTCGCAAGGAACTGCAGGATGCTGGAGCGCATGGACTTAGAAGAGTGTGTCCTTATCACCGACGCCACCCTGGTGCACCTGTCCATGGGCTGTCCGCGACTTGAGAAACTGACGCTGTCCCACTGTGACCTGATCACGGACAACGGCATAAAGCAGCTGTCCCTGTCCCCCTGCGCCGCCGAGCACCTCACCGTGCTGGGTCTGGACAACTGTCCCCTGGTGACCGACGAGGCGCTGGAGCACCTGACGTCCTGCCACAACCTGCAGCTCATCGAGCTCTACGACTGCCAGATGGTCACGAGGAACGCCATACGGAAGCTCAGGAACCATCTTCCCAACATCAAAGTGCACGCGTACTTCGCCCCGGTGACTCCGCCCGCGGCCGCCGGCGGCGCCCGGCCCCGCTACTGCCGCTGCTGCAACATCATATGA
Protein
MTHSGRSKFEITRFTNEDENHINRKLPKELLLRILSYLDVVSLCRCAQVSKLWNILALDGSNWQRIDLFEFQRDVEGPVIENISQRCGGFLRQLSLRGCESIGDGSMKTLAQSCPNIEDLNLNKCKKITDQTCQALGRRCSKLQKINLDSCPSVSDISLKALADGCPLLTHVNVSWCQSITESGVEALARGCPKLKSFICKGCKNVNDRAVISLATRCPDLEVLDIQGCETVSDEAVSRLGGALRRLCASGCARLTDASLAALAAAAPRLATLELAQCTQLTDAGFQALARNCRMLERMDLEECVLITDATLVHLSMGCPRLEKLTLSHCDLITDNGIKQLSLSPCAAEHLTVLGLDNCPLVTDEALEHLTSCHNLQLIELYDCQMVTRNAIRKLRNHLPNIKVHAYFAPVTPPAAAGGARPRYCRCCNII
Summary
Uniprot
A0A1E1WFQ2
A0A194PT08
A0A212EKX3
A0A194QZH6
A0A2A4K2D2
A0A1Y1LT38
+ More
A0A1Y1LV50 A0A2P8XL67 A0A139WHE4 A0A1B6MJR5 D6WMF2 A0A1B6DH79 A0A1B6EUJ6 A0A146LWP7 A0A1B6KVT6 K7INM3 A0A0C9QX29 A0A023F5G5 A0A0V0GAG9 A0A2H1VZD7 T1H911 A0A0A9Y771 A0A1B6EU84 A0A0A9YA79 A0A0P4VW86 A0A158N964 A0A2A3E8A6 A0A154P5E3 A0A195B4F6 E2C4H9 A0A3L8E484 A0A195CTX1 F4WIT3 A0A195FPD9 A0A151WLY0 A0A026VXP2 A0A088AGT2 A0A1B6LKJ7 U5ELL2 A0A084VDS1 A0A0L7R6S6 A0A182RVM9 A0A1Q3F5N1 A0A182TE07 A0A1S4GWA2 A0A182X1J9 A0A182L2H4 A0A182IEI0 B0WXG5 A0A182MZC9 A0A2M4BMN0 Q172D0 A0A182JZ35 A0A182XZL2 A0A0C9RSD8 A0A2M4AI82 A0A2M4CTS8 A0A182SD55 A0A1B6MJZ4 A0A182W7S5 A0A182P0E5 A0A182FN71 A0A2M3Z778 Q7Q3C3 A0A1B6JLU7 A0A182J008 A0A182M899 W5JVH0 A0A2S2P0S8 A0A1L8DAU2 J9JNE9 A0A0P5ZA17 A0A2H8TJB7 A0A232F4P2 A0A2S2PWU0 A0A182H6A0 E9G1Z9 E0VCM3 A0A1D2N5M0 A0A151IWR0 A0A1B6JY84 A0A3B0JUE8 B4LPX8 A0A182Q639 A0A0P6E6D8 A0A0J7L1K5 A0A0R3NS81 Q5BI22 A0A0B4KEK4 A0A0B4KEU4 A0A1W4UID8 Q291I1 A0A0J9RAF5 A0A0R1DQF0 A0A0Q5W2A3 A8DYA5 B4GAP3 A0A0J9RAI0 E2ADF4 A1Z8M8
A0A1Y1LV50 A0A2P8XL67 A0A139WHE4 A0A1B6MJR5 D6WMF2 A0A1B6DH79 A0A1B6EUJ6 A0A146LWP7 A0A1B6KVT6 K7INM3 A0A0C9QX29 A0A023F5G5 A0A0V0GAG9 A0A2H1VZD7 T1H911 A0A0A9Y771 A0A1B6EU84 A0A0A9YA79 A0A0P4VW86 A0A158N964 A0A2A3E8A6 A0A154P5E3 A0A195B4F6 E2C4H9 A0A3L8E484 A0A195CTX1 F4WIT3 A0A195FPD9 A0A151WLY0 A0A026VXP2 A0A088AGT2 A0A1B6LKJ7 U5ELL2 A0A084VDS1 A0A0L7R6S6 A0A182RVM9 A0A1Q3F5N1 A0A182TE07 A0A1S4GWA2 A0A182X1J9 A0A182L2H4 A0A182IEI0 B0WXG5 A0A182MZC9 A0A2M4BMN0 Q172D0 A0A182JZ35 A0A182XZL2 A0A0C9RSD8 A0A2M4AI82 A0A2M4CTS8 A0A182SD55 A0A1B6MJZ4 A0A182W7S5 A0A182P0E5 A0A182FN71 A0A2M3Z778 Q7Q3C3 A0A1B6JLU7 A0A182J008 A0A182M899 W5JVH0 A0A2S2P0S8 A0A1L8DAU2 J9JNE9 A0A0P5ZA17 A0A2H8TJB7 A0A232F4P2 A0A2S2PWU0 A0A182H6A0 E9G1Z9 E0VCM3 A0A1D2N5M0 A0A151IWR0 A0A1B6JY84 A0A3B0JUE8 B4LPX8 A0A182Q639 A0A0P6E6D8 A0A0J7L1K5 A0A0R3NS81 Q5BI22 A0A0B4KEK4 A0A0B4KEU4 A0A1W4UID8 Q291I1 A0A0J9RAF5 A0A0R1DQF0 A0A0Q5W2A3 A8DYA5 B4GAP3 A0A0J9RAI0 E2ADF4 A1Z8M8
Pubmed
26354079
22118469
28004739
29403074
18362917
19820115
+ More
26823975 20075255 25474469 25401762 27129103 21347285 20798317 30249741 21719571 24508170 24438588 12364791 20966253 17510324 25244985 20920257 23761445 28648823 26483478 21292972 20566863 27289101 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304
26823975 20075255 25474469 25401762 27129103 21347285 20798317 30249741 21719571 24508170 24438588 12364791 20966253 17510324 25244985 20920257 23761445 28648823 26483478 21292972 20566863 27289101 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304
EMBL
GDQN01005224
JAT85830.1
KQ459593
KPI96447.1
AGBW02014188
OWR42129.1
+ More
KQ460890 KPJ10857.1 NWSH01000243 PCG78064.1 GEZM01049038 JAV76151.1 GEZM01049039 JAV76150.1 PYGN01001799 PSN32749.1 KQ971343 KYB27304.1 GEBQ01003789 JAT36188.1 EFA03310.2 GEDC01012264 JAS25034.1 GECZ01028198 GECZ01019545 GECZ01019168 GECZ01000074 JAS41571.1 JAS50224.1 JAS50601.1 JAS69695.1 GDHC01007194 JAQ11435.1 GEBQ01024408 JAT15569.1 GBYB01000240 JAG70007.1 GBBI01002324 JAC16388.1 GECL01001002 JAP05122.1 ODYU01005396 SOQ46188.1 ACPB03019912 GBHO01015600 GBHO01015598 GBRD01015499 GDHC01006679 JAG28004.1 JAG28006.1 JAG50327.1 JAQ11950.1 GECZ01030645 GECZ01028214 GECZ01008182 JAS39124.1 JAS41555.1 JAS61587.1 GBHO01015601 GBHO01015596 GBRD01015498 GDHC01002683 GDHC01000373 JAG28003.1 JAG28008.1 JAG50328.1 JAQ15946.1 JAQ18256.1 GDKW01001641 JAI54954.1 ADTU01008734 KZ288326 PBC27987.1 KQ434822 KZC07072.1 KQ976618 KYM79152.1 GL452471 EFN77163.1 QOIP01000001 RLU27352.1 KQ977279 KYN04136.1 GL888177 EGI65879.1 KQ981382 KYN42261.1 KQ982959 KYQ48705.1 KK107602 EZA48578.1 GEBQ01015745 JAT24232.1 GANO01001355 JAB58516.1 ATLV01011730 ATLV01011731 KE524715 KFB36115.1 KQ414646 KOC66529.1 GFDL01012232 JAV22813.1 AAAB01008964 APCN01005204 DS232164 EDS36495.1 GGFJ01005198 MBW54339.1 CH477441 EAT40856.1 GBYB01010316 JAG80083.1 GGFK01007178 MBW40499.1 GGFL01004403 MBW68581.1 GEBQ01003754 JAT36223.1 GGFM01003547 MBW24298.1 EAA12920.4 GECU01007634 JAT00073.1 AXCM01014285 ADMH02000216 ETN67343.1 GGMR01010381 MBY23000.1 GFDF01010505 JAV03579.1 ABLF02011771 GDIQ01150435 GDIP01048958 LRGB01000032 JAL01291.1 JAM54757.1 KZS21231.1 GFXV01002438 MBW14243.1 NNAY01000942 OXU25806.1 GGMS01000804 MBY70007.1 JXUM01113704 KQ565716 KXJ70748.1 GL732529 EFX86579.1 DS235059 EEB11129.1 LJIJ01000198 ODN00568.1 KQ980846 KYN12261.1 GECU01003557 JAT04150.1 OUUW01000001 SPP74698.1 CH940648 EDW61318.2 AXCN02000604 GDIP01046022 GDIQ01068466 JAM57693.1 JAN26271.1 LBMM01001343 KMQ96478.1 CM000071 KRT01869.1 BT021402 AE013599 AAX33550.1 ABV53766.1 AGB93433.1 AGB93434.1 EAL25131.3 CM002911 KMY93016.1 CM000158 KRJ99509.1 CH954179 KQS62872.1 BT120055 ABV53765.1 ADA53594.1 CH479181 EDW31995.1 KMY93017.1 GL438750 EFN68516.1 AAF58635.1
KQ460890 KPJ10857.1 NWSH01000243 PCG78064.1 GEZM01049038 JAV76151.1 GEZM01049039 JAV76150.1 PYGN01001799 PSN32749.1 KQ971343 KYB27304.1 GEBQ01003789 JAT36188.1 EFA03310.2 GEDC01012264 JAS25034.1 GECZ01028198 GECZ01019545 GECZ01019168 GECZ01000074 JAS41571.1 JAS50224.1 JAS50601.1 JAS69695.1 GDHC01007194 JAQ11435.1 GEBQ01024408 JAT15569.1 GBYB01000240 JAG70007.1 GBBI01002324 JAC16388.1 GECL01001002 JAP05122.1 ODYU01005396 SOQ46188.1 ACPB03019912 GBHO01015600 GBHO01015598 GBRD01015499 GDHC01006679 JAG28004.1 JAG28006.1 JAG50327.1 JAQ11950.1 GECZ01030645 GECZ01028214 GECZ01008182 JAS39124.1 JAS41555.1 JAS61587.1 GBHO01015601 GBHO01015596 GBRD01015498 GDHC01002683 GDHC01000373 JAG28003.1 JAG28008.1 JAG50328.1 JAQ15946.1 JAQ18256.1 GDKW01001641 JAI54954.1 ADTU01008734 KZ288326 PBC27987.1 KQ434822 KZC07072.1 KQ976618 KYM79152.1 GL452471 EFN77163.1 QOIP01000001 RLU27352.1 KQ977279 KYN04136.1 GL888177 EGI65879.1 KQ981382 KYN42261.1 KQ982959 KYQ48705.1 KK107602 EZA48578.1 GEBQ01015745 JAT24232.1 GANO01001355 JAB58516.1 ATLV01011730 ATLV01011731 KE524715 KFB36115.1 KQ414646 KOC66529.1 GFDL01012232 JAV22813.1 AAAB01008964 APCN01005204 DS232164 EDS36495.1 GGFJ01005198 MBW54339.1 CH477441 EAT40856.1 GBYB01010316 JAG80083.1 GGFK01007178 MBW40499.1 GGFL01004403 MBW68581.1 GEBQ01003754 JAT36223.1 GGFM01003547 MBW24298.1 EAA12920.4 GECU01007634 JAT00073.1 AXCM01014285 ADMH02000216 ETN67343.1 GGMR01010381 MBY23000.1 GFDF01010505 JAV03579.1 ABLF02011771 GDIQ01150435 GDIP01048958 LRGB01000032 JAL01291.1 JAM54757.1 KZS21231.1 GFXV01002438 MBW14243.1 NNAY01000942 OXU25806.1 GGMS01000804 MBY70007.1 JXUM01113704 KQ565716 KXJ70748.1 GL732529 EFX86579.1 DS235059 EEB11129.1 LJIJ01000198 ODN00568.1 KQ980846 KYN12261.1 GECU01003557 JAT04150.1 OUUW01000001 SPP74698.1 CH940648 EDW61318.2 AXCN02000604 GDIP01046022 GDIQ01068466 JAM57693.1 JAN26271.1 LBMM01001343 KMQ96478.1 CM000071 KRT01869.1 BT021402 AE013599 AAX33550.1 ABV53766.1 AGB93433.1 AGB93434.1 EAL25131.3 CM002911 KMY93016.1 CM000158 KRJ99509.1 CH954179 KQS62872.1 BT120055 ABV53765.1 ADA53594.1 CH479181 EDW31995.1 KMY93017.1 GL438750 EFN68516.1 AAF58635.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000218220
UP000245037
UP000007266
+ More
UP000002358 UP000015103 UP000005205 UP000242457 UP000076502 UP000078540 UP000008237 UP000279307 UP000078542 UP000007755 UP000078541 UP000075809 UP000053097 UP000005203 UP000030765 UP000053825 UP000075900 UP000075902 UP000076407 UP000075882 UP000075840 UP000002320 UP000075884 UP000008820 UP000075881 UP000076408 UP000075901 UP000075920 UP000075885 UP000069272 UP000007062 UP000075880 UP000075883 UP000000673 UP000007819 UP000076858 UP000215335 UP000069940 UP000249989 UP000000305 UP000009046 UP000094527 UP000078492 UP000268350 UP000008792 UP000075886 UP000036403 UP000001819 UP000000803 UP000192221 UP000002282 UP000008711 UP000008744 UP000000311
UP000002358 UP000015103 UP000005205 UP000242457 UP000076502 UP000078540 UP000008237 UP000279307 UP000078542 UP000007755 UP000078541 UP000075809 UP000053097 UP000005203 UP000030765 UP000053825 UP000075900 UP000075902 UP000076407 UP000075882 UP000075840 UP000002320 UP000075884 UP000008820 UP000075881 UP000076408 UP000075901 UP000075920 UP000075885 UP000069272 UP000007062 UP000075880 UP000075883 UP000000673 UP000007819 UP000076858 UP000215335 UP000069940 UP000249989 UP000000305 UP000009046 UP000094527 UP000078492 UP000268350 UP000008792 UP000075886 UP000036403 UP000001819 UP000000803 UP000192221 UP000002282 UP000008711 UP000008744 UP000000311
Interpro
Gene 3D
ProteinModelPortal
A0A1E1WFQ2
A0A194PT08
A0A212EKX3
A0A194QZH6
A0A2A4K2D2
A0A1Y1LT38
+ More
A0A1Y1LV50 A0A2P8XL67 A0A139WHE4 A0A1B6MJR5 D6WMF2 A0A1B6DH79 A0A1B6EUJ6 A0A146LWP7 A0A1B6KVT6 K7INM3 A0A0C9QX29 A0A023F5G5 A0A0V0GAG9 A0A2H1VZD7 T1H911 A0A0A9Y771 A0A1B6EU84 A0A0A9YA79 A0A0P4VW86 A0A158N964 A0A2A3E8A6 A0A154P5E3 A0A195B4F6 E2C4H9 A0A3L8E484 A0A195CTX1 F4WIT3 A0A195FPD9 A0A151WLY0 A0A026VXP2 A0A088AGT2 A0A1B6LKJ7 U5ELL2 A0A084VDS1 A0A0L7R6S6 A0A182RVM9 A0A1Q3F5N1 A0A182TE07 A0A1S4GWA2 A0A182X1J9 A0A182L2H4 A0A182IEI0 B0WXG5 A0A182MZC9 A0A2M4BMN0 Q172D0 A0A182JZ35 A0A182XZL2 A0A0C9RSD8 A0A2M4AI82 A0A2M4CTS8 A0A182SD55 A0A1B6MJZ4 A0A182W7S5 A0A182P0E5 A0A182FN71 A0A2M3Z778 Q7Q3C3 A0A1B6JLU7 A0A182J008 A0A182M899 W5JVH0 A0A2S2P0S8 A0A1L8DAU2 J9JNE9 A0A0P5ZA17 A0A2H8TJB7 A0A232F4P2 A0A2S2PWU0 A0A182H6A0 E9G1Z9 E0VCM3 A0A1D2N5M0 A0A151IWR0 A0A1B6JY84 A0A3B0JUE8 B4LPX8 A0A182Q639 A0A0P6E6D8 A0A0J7L1K5 A0A0R3NS81 Q5BI22 A0A0B4KEK4 A0A0B4KEU4 A0A1W4UID8 Q291I1 A0A0J9RAF5 A0A0R1DQF0 A0A0Q5W2A3 A8DYA5 B4GAP3 A0A0J9RAI0 E2ADF4 A1Z8M8
A0A1Y1LV50 A0A2P8XL67 A0A139WHE4 A0A1B6MJR5 D6WMF2 A0A1B6DH79 A0A1B6EUJ6 A0A146LWP7 A0A1B6KVT6 K7INM3 A0A0C9QX29 A0A023F5G5 A0A0V0GAG9 A0A2H1VZD7 T1H911 A0A0A9Y771 A0A1B6EU84 A0A0A9YA79 A0A0P4VW86 A0A158N964 A0A2A3E8A6 A0A154P5E3 A0A195B4F6 E2C4H9 A0A3L8E484 A0A195CTX1 F4WIT3 A0A195FPD9 A0A151WLY0 A0A026VXP2 A0A088AGT2 A0A1B6LKJ7 U5ELL2 A0A084VDS1 A0A0L7R6S6 A0A182RVM9 A0A1Q3F5N1 A0A182TE07 A0A1S4GWA2 A0A182X1J9 A0A182L2H4 A0A182IEI0 B0WXG5 A0A182MZC9 A0A2M4BMN0 Q172D0 A0A182JZ35 A0A182XZL2 A0A0C9RSD8 A0A2M4AI82 A0A2M4CTS8 A0A182SD55 A0A1B6MJZ4 A0A182W7S5 A0A182P0E5 A0A182FN71 A0A2M3Z778 Q7Q3C3 A0A1B6JLU7 A0A182J008 A0A182M899 W5JVH0 A0A2S2P0S8 A0A1L8DAU2 J9JNE9 A0A0P5ZA17 A0A2H8TJB7 A0A232F4P2 A0A2S2PWU0 A0A182H6A0 E9G1Z9 E0VCM3 A0A1D2N5M0 A0A151IWR0 A0A1B6JY84 A0A3B0JUE8 B4LPX8 A0A182Q639 A0A0P6E6D8 A0A0J7L1K5 A0A0R3NS81 Q5BI22 A0A0B4KEK4 A0A0B4KEU4 A0A1W4UID8 Q291I1 A0A0J9RAF5 A0A0R1DQF0 A0A0Q5W2A3 A8DYA5 B4GAP3 A0A0J9RAI0 E2ADF4 A1Z8M8
PDB
1FS2
E-value=1.31547e-11,
Score=168
Ontologies
KEGG
GO
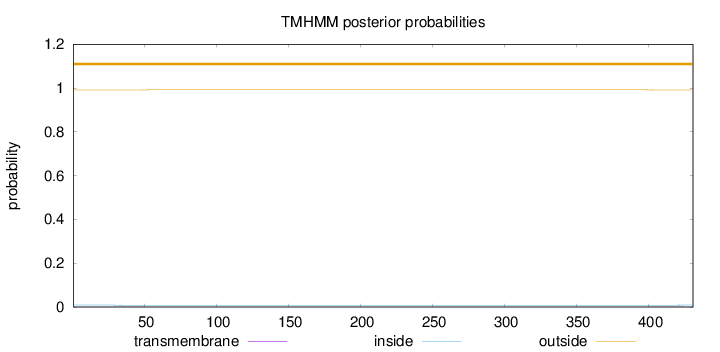
Topology
Length:
431
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02885
Exp number, first 60 AAs:
0.01308
Total prob of N-in:
0.00893
outside
1 - 431
Population Genetic Test Statistics
Pi
242.398301
Theta
196.291403
Tajima's D
1.837089
CLR
0.02305
CSRT
0.857607119644018
Interpretation
Uncertain
