Gene
KWMTBOMO00993
Pre Gene Modal
BGIBMGA001376
Annotation
PREDICTED:_protein_quiver_[Bombyx_mori]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 3.161
Sequence
CDS
ATGCGCTGGATATTTCCCGTTCCAGTTCTCCTCATGTCCTGGTTCGGTGTGGACCAAGTTAAAGGGAATTCCTGCATCAGAACTGCCCGCACGATATACTGTTACGAGTGTGACAGCTGGTCGGACCCGCGATGCAAGGACCCGTTCAACTACACGGCGCTGCCGCGGGACCAGCCCCCGCTCCAGACCTGTAACGGGTGCTGCGTCAAAATGGTGCGCTATTCCAAAAGCCCTTACGAGGTCGTCAGACGAACGTGCACTTCTCAACTACAAATAAACTTGTTCATGGTGGACCACGTGTGCATGATGGAAAGTACTGGCACTGGTCACATGTGTTTCTGTGAGGAGGACATGTGTAACGGGGCGCCGGCCAGGACGGCCGCAGTCCTGGCCGTTGTCGCGGCGCTAGCGCTCGCATGCTGCATGAGACGGGCCGTCTGCATGCCTCGGGTACAGGAGTGCGGCCGCCGCGGCTCGCCACAACTTAGGTTAGCTACGTGGATATTCTACGGTGTACCATAG
Protein
MRWIFPVPVLLMSWFGVDQVKGNSCIRTARTIYCYECDSWSDPRCKDPFNYTALPRDQPPLQTCNGCCVKMVRYSKSPYEVVRRTCTSQLQINLFMVDHVCMMESTGTGHMCFCEEDMCNGAPARTAAVLAVVAALALACCMRRAVCMPRVQECGRRGSPQLRLATWIFYGVP
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
EMBL
Proteomes
Pfam
PF17064 QVR
Interpro
IPR031424
QVR
ProteinModelPortal
Ontologies
GO
Topology
Subcellular location
Cell membrane
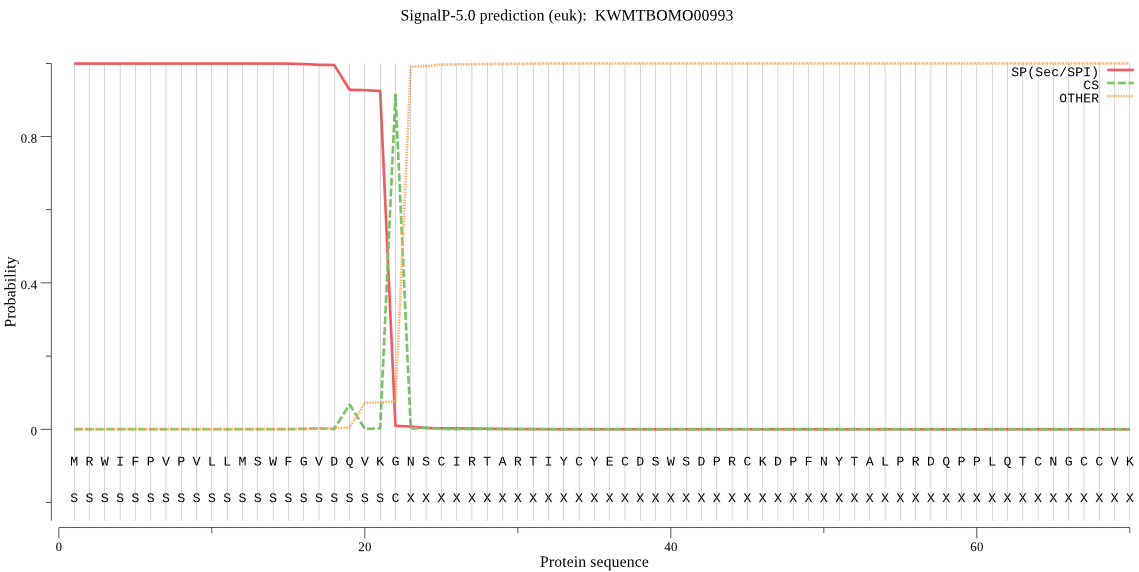
SignalP
Position: 1 - 22,
Likelihood: 0.999440
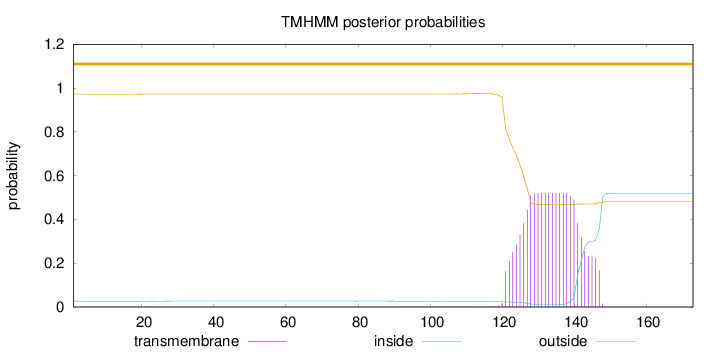
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.72785
Exp number, first 60 AAs:
0.0574
Total prob of N-in:
0.02678
outside
1 - 173
Population Genetic Test Statistics
Pi
226.805588
Theta
150.075962
Tajima's D
1.527587
CLR
0.19135
CSRT
0.794510274486276
Interpretation
Uncertain
