Gene
KWMTBOMO00988 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001370
Annotation
Talin-1_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.152
Sequence
CDS
ATGGCGACCCTATCGTTGAAGATCTCCATTGACGGAGGCAAGGTCGTGAAGACAATACAGTTCGAACCCTCCACCACAGTGTACGACGCGTGCCGCATCATCAGGGAAAAGATCTTGGAGGCCACAGCGGGAGACCCAAAAGAGTATGGCCTCTTCTTGGCTTCGGAGGAAGACAACAAGAAGGGGATATGGCTGGAAGCGTCCCGAAGTCTTGACTATTACATGCTAAGGAACGGAGACCTTCTGGAGTACAACAAAAAGACCAGGAATCTCAGAGTCAGGATGCTTGATGGTACTGTGAAAACCCTGCTGGTGGACGACAGTCAGATCGTCGCGAACCTCATGGTAGTGATTTGCACCAAGATCGGGATCACCAACTATGACGAATACGGCCTGGTTCGGGAAGAGCAAAAGGAGGAACCGGATCCCTGCGAGAAGCCAAACTACGGGACTCTGACCCTGAAGCGTAAGCACCAGGAGAAGGAACGAGACGCCAAGATGGAACAGCTCAGGAAGAAACTCAGAACTGATGACGAAGTGAACTGGGTGGAGGGGTCGCTGACCCTTCGTGAGCAGGGCGTGGAGGCGGGCGAGACCTTACTGCTGCGCCGCAAGCTGTTCTACTCGGACAGGAACGTGGACTCGCGCGACCCCGTGCAGCTCACCCTGCTGTACGTGCAGGCCAGGGACGCCATCCTGGCCGGCGCGCACCCCACCACGCAGGAGAAAGCTTGCGAGTTTGCCGGGATACAGTGCCAGATACAGTTCGGGGACCACCGGGAGGACAAGCACACGCCAGGATTCCTCGATTTAAAAGAGTTCCTGCCGGCCTCGTACGTGAAGGTGAAAGGCATCGAGAAGAAGGTGTTCAGGGAGCACAAGAAGCACGTCGGGCTCAGCGAGCTGGACGCCAAGGTGCTCTACACCAAGACGGCGCGGGACCTCAAGACCTACGGAGTCGCCTTCTTCCTCGTCAAGGAAAAAATGAAAGGCAAAAACAAACTGGTACCGCGTCTTCTGGGAGTCACGAAGGACTCCGTGCTCAGACTGGACGAGAAGACGAAGGAGATCCTGCAGACGTGGCCGCTCACCACCGTGAGGAGGTGGTGCGCCAGCCCCAACACTTTCACCCTCGACTTTGGCGATTACAGCGACCAGTACTACTCGGTGCAGACCACGGAGGCGGAGCAGATCCTGCAGGTCATCGCCGGCTACATCGACATCATCGTGCGGAAGCAGCGGGCCAAGGAGCACCTCGGCATCGAGGGAGACGAGGGCTCCGCCATGCTCGAGGACTCCGTGTCGCCCTCCAAAGCGAACATCATCCAGCACGACACGTTCAAGTCCGGTAAAGTGAACACCGAGTCCGTCGCCAAGCCGGCGGTGCACCGGCCCGGAGCTGAAGGTGCGAAGCCGTTCGCCGTGGGGCACGTGACGGGCGCGCAGCTGACCACGGTCTCCGGGCAGGTGGTGACGGGGCACGCGCCGCCCGCCGCGTCGCAGGTACAGCAGACCAAGGTGACCTCGATACTCACGGAACCTCAGCGGGCCCTGTTGACCACCATCACCAGCGGCAGGGAGATCATCAAGCAGACCGAAGAAGGACTCAGCAGGACAACCCTGCCCGAACTGGGAGGTGACGCATCCTCAGTGAAATGGAAGCAGTCGACCCTGGACACCAACAAGCAGGCCGTCAGCTCGCAGATCGCGGCCATGAACGCGGCCACGGCGCAGGTCGTCACCCTCACCGCCGGTCCAACCGAAGAGGTAGACCACACAGCAGTGGGTGCCGCCATCACGACCATCACCAGCAACCTGCCGGAAGTGACGCGCGGGGTCAGGATGATCGCAGCCCTCATGGACGACTCCGACAACGGAGACAGACTGTTGGACGCCACCAGGAAACTTTGCACCGCGTTCACCGACCTGCTGAAGGCGGCCGAGCCGGAAACGAAGGAGCCGCGCCAGAACCTGTTGAACGCAGCCTCCAGGGTCGGGGAGGCGTCCACGTCCGTGCTGTACACCATCGGGGAGGAGACCGAGGAGGACAAGCAGACGCAGGTAACCGAGCCGGATAAAAAGAAAAAATCCTTCCTCTACAGGAAACTCAATTACTGCTACGACAACTTCTACAACGTCTTCTTTAGTAAGGACAAGTTCGGAGATGATTTCGACGATGGTAGAGTAGACGACGGCGTGTATGAGGACGTCGACGTCACCAGGACCTGGGGCCTGGACGCCATCCAGGAGGAATCCGATAGTGACTACTACAGAAGCCTGGCCTACTGCGACGCCGTGCTACGCCCCGGAAGGCCGGCCAACTTCCACAACAACCGCACTGATGACGCGGACCTAGAGTCCATCATAGGGACGTGCGTCGATTATCTCGATAAGAAAATCGATAACAAAACCGAATTGAATAACAGCGACAGCGATTACGAACCGATAGGCTTCGCGCCCGGCCGCCCCCCGCGACCCCCGCCGACGGTCACGGACAAGATATACAACTTCAAAAATGATTTTTCTAATCACGACTACGCTAACGTAAACTACGACCCGCCGAAACACGACTACTCCAGCGTCAACCTCCACCGCCGGACCAAAAACGATATATTGAGGCAACGGTTCTTTCTATCGAACAATATCGATAAAAACGATATAAACGATCGATTCAATCGATACAAGGATAAGAAGATCGAATGTCACAGCATCAGGAACGACGTCACCAAATCCGGTCCCGACGTGAGCGTGGCCGGCGCTCTGAACCTCAAAATCCTAAACCCGGATAGATACAACCGGGTGCTGACGACGAAGGAGACGTTCTACGAGGAATCCGAAGCCAGCTTCACTAAGACGAGCCTATCGAAACTAAAGAACGATCGCTTCGAGAATTTTGTTAAGAAAAGCGAGAAGTACTACACTAGTAGTGACGGTAGCGACCGCAAAACAGAAATCTTAAATAATTCCGAGGGACAACTTAGACCCAAAACGGAGATACGCATCGTGTACAACCCGACGTTCCGCGTGCGGGGCGGACACGACCGCGGCGCCCGCCCGTGCTCGTCTCCCAGATTTCACCTCATGATCCTGTCATGGGTGATGTCTGGATTGATTAATATTATTATAGGGTTGATGGACATCCTTCTGTCTCTCGCCAAAGCCGTAGCCAACACGACGGCAGCTTTAGTGTTGAAAGCGAAGAACGTGGCGGCCCAGTGCCGGGACGACCAGCAGCTGCAGAACAACATCATAGCCGCGGCCACGCACTGCGCCCTGGCCACCAGCCAGCTCGTGGCCTGCGCCAAGGTGGTGGCGCCCACCCTGCACAGCGCGGCGTGTCGCGAGCAGCTGACGAGCGCGGCGCGGCAAGTGGCGCAGGCCGTGGACCGGCTGGTGGAGGCCTGCGCGCCCGCACCGGCCGCGGCGGCGCCCGCGCGCGACCAGCTCGCCGCCGCCGCGCACTGCGTCTCCGAGGAGCTGGAGAGGCTGCTGGCCCACTGTCAACTTGAGCGCGGGGGCGCGGCGGCGGCGGCGGGGCGCGCGGTGGAGGCGGTGCTGGGCGCGCACGAGCGGCTGCTGGCGGCCGCCGGGCCAGCCGACATGCTGCGCCACGCCAGGCAGCTGGGACACGCCACGGCGCTGCTCATCACCAACATCAAGACCGAGGCCGAGGAGTCACCGTCGGACTCCCAGCGCAAGCTCCTAGCCGCCGCCAAGCTGCTGGCCGACGCCACGGCCCGCATGGTGGAGGCGGCCCGCCAGTGCGCCTCCTCGCCACAGGATCGGGAGAAGCAAGAGGTGCTCCGCAAAGCGGCGGAGGAGCTGCGCTTCATCACGGCCGACTACGCGCAGGGACAGGACATCGTCGGCTCACAAGTGGCCAGGCTGCAGGAAAGCGCCCGGCAGGCGACGTCCAGCGCCACGCAGCTGATAACGTCGGCCCAGAACGCAACGCAGTACAACACCAACAGATACTCTCAGGAGACCCTTCTAGAAGAGTGCAAAGTGCTGAACGAGCACCTCCCCCGCATGGTGGAAGCCGTGAACGTGTCCAACGTGAAGCCGACGCAGGCAGCGGCCAGGCTCGACCTCATCTCTGCATCTGAAGCCTTCTTACAGCCAAGTGGCCACGTGATCTCAGCAGCCCGAGGCGCCCTGCCGACCGTAAGCGAGCCCTCCGCAGCCAAGCACCTGGCCGACAACACGCAGCTCTACACTTCAAGCTGCGCCGACCTCAGGTCCGCAGTGAGCAGAGCCAGGATATCCTGCAAGGGCTTGGAGCTGGACGCAGCGGCGGAGATCATCAAATCCCTGCAAGAGGAGCTTGATGAGCTGGAGGAGGCGGTCAGGGCTTTCGACCTGAAACCCTTACCCGGGCAGACGGCGGAGTGGGCCGCGGCCACCATGAACTCCGCGTCCCGCGCCGCCGCCAGCTCCACCGCGCGCCTCGTGTCCGGCGCCAGCGGCTCCCACGCCGCGCCCGACCTCGCGCACGCGCTGCGGGACTTCACGCACGGGCTGCGCGCCGCCGCCGCCGCCGCGCCGCCTCCCCGCGCGCACAGAGTGATATCATCAGGGCGGCAGGTCCTGTACTCGTCCCTGACCCTCGTGGAGACGGTCAAGAAGCAGATCAGCAGCTCGGAGCAGGTGGACAGCGCCGCGCTCGCCTCCATAGCCAAGGACATCACGCACGGTCTAGAGCAGACGATGGAAGCGGTTCCGTCTCACACAGAGCTGGACCTCGCCATGGAGAACATCAACGAGACCCTCAACATTCTGAACATGGGCGAGTTCCCGCCCACGGACAGCTCCTATGGGGAACTCCAAGTCCAGCTGAACTCGGCGGCAGCAGGCCTCAGCCAGGCCTCGTCGGCCGTGGTGCAGAGCGTGGACGACCCCCGGACCCTGGCGGACTCCAGCCACAGCTTCGGACAGGCCTTCAACAATCTGCTCGGAGTCGGCATGGAGATGGCTGGACAAACTGAGGACCGCGAGACTCAGTCGTTGATGGTGAACTCCATGAAGTCGGTGACCGTCAACTCCTCCAAGCTCCTCGGGACAGCGAAGTCCGTCAGCCAGGACTTATCCCGCCCGAACGCCAAGAACCAGCTGACGGCCGCCGCCCGCGCCGTCACCGACAGCATCAACAAGCTGGTGGACGCGTGCACGCAGGCGGCGCCGGGACACAAGGCGTGCGGCGCGGCGCGGCGCAGCGTGCAGGCCGCGCGCCCGCTGCTGGACGCGCCCGCGCAGCCGCTCTCCGACCTCTCCTACTTCGCGTGTCTGGACGCCGTGCTCGACCACAGCAAGCAGCTCAGTGAGTGA
Protein
MATLSLKISIDGGKVVKTIQFEPSTTVYDACRIIREKILEATAGDPKEYGLFLASEEDNKKGIWLEASRSLDYYMLRNGDLLEYNKKTRNLRVRMLDGTVKTLLVDDSQIVANLMVVICTKIGITNYDEYGLVREEQKEEPDPCEKPNYGTLTLKRKHQEKERDAKMEQLRKKLRTDDEVNWVEGSLTLREQGVEAGETLLLRRKLFYSDRNVDSRDPVQLTLLYVQARDAILAGAHPTTQEKACEFAGIQCQIQFGDHREDKHTPGFLDLKEFLPASYVKVKGIEKKVFREHKKHVGLSELDAKVLYTKTARDLKTYGVAFFLVKEKMKGKNKLVPRLLGVTKDSVLRLDEKTKEILQTWPLTTVRRWCASPNTFTLDFGDYSDQYYSVQTTEAEQILQVIAGYIDIIVRKQRAKEHLGIEGDEGSAMLEDSVSPSKANIIQHDTFKSGKVNTESVAKPAVHRPGAEGAKPFAVGHVTGAQLTTVSGQVVTGHAPPAASQVQQTKVTSILTEPQRALLTTITSGREIIKQTEEGLSRTTLPELGGDASSVKWKQSTLDTNKQAVSSQIAAMNAATAQVVTLTAGPTEEVDHTAVGAAITTITSNLPEVTRGVRMIAALMDDSDNGDRLLDATRKLCTAFTDLLKAAEPETKEPRQNLLNAASRVGEASTSVLYTIGEETEEDKQTQVTEPDKKKKSFLYRKLNYCYDNFYNVFFSKDKFGDDFDDGRVDDGVYEDVDVTRTWGLDAIQEESDSDYYRSLAYCDAVLRPGRPANFHNNRTDDADLESIIGTCVDYLDKKIDNKTELNNSDSDYEPIGFAPGRPPRPPPTVTDKIYNFKNDFSNHDYANVNYDPPKHDYSSVNLHRRTKNDILRQRFFLSNNIDKNDINDRFNRYKDKKIECHSIRNDVTKSGPDVSVAGALNLKILNPDRYNRVLTTKETFYEESEASFTKTSLSKLKNDRFENFVKKSEKYYTSSDGSDRKTEILNNSEGQLRPKTEIRIVYNPTFRVRGGHDRGARPCSSPRFHLMILSWVMSGLINIIIGLMDILLSLAKAVANTTAALVLKAKNVAAQCRDDQQLQNNIIAAATHCALATSQLVACAKVVAPTLHSAACREQLTSAARQVAQAVDRLVEACAPAPAAAAPARDQLAAAAHCVSEELERLLAHCQLERGGAAAAAGRAVEAVLGAHERLLAAAGPADMLRHARQLGHATALLITNIKTEAEESPSDSQRKLLAAAKLLADATARMVEAARQCASSPQDREKQEVLRKAAEELRFITADYAQGQDIVGSQVARLQESARQATSSATQLITSAQNATQYNTNRYSQETLLEECKVLNEHLPRMVEAVNVSNVKPTQAAARLDLISASEAFLQPSGHVISAARGALPTVSEPSAAKHLADNTQLYTSSCADLRSAVSRARISCKGLELDAAAEIIKSLQEELDELEEAVRAFDLKPLPGQTAEWAAATMNSASRAAASSTARLVSGASGSHAAPDLAHALRDFTHGLRAAAAAAPPPRAHRVISSGRQVLYSSLTLVETVKKQISSSEQVDSAALASIAKDITHGLEQTMEAVPSHTELDLAMENINETLNILNMGEFPPTDSSYGELQVQLNSAAAGLSQASSAVVQSVDDPRTLADSSHSFGQAFNNLLGVGMEMAGQTEDRETQSLMVNSMKSVTVNSSKLLGTAKSVSQDLSRPNAKNQLTAAARAVTDSINKLVDACTQAAPGHKACGAARRSVQAARPLLDAPAQPLSDLSYFACLDAVLDHSKQLSE
Summary
Uniprot
Pubmed
EMBL
Proteomes
Pfam
Interpro
IPR035964
I/LWEQ_dom_sf
+ More
IPR000299 FERM_domain
IPR015009 Vinculin-bd_dom
IPR015224 Talin_cent
IPR019748 FERM_central
IPR036723 Alpha-catenin/vinculin-like_sf
IPR030224 Sla2_fam
IPR011993 PH-like_dom_sf
IPR036476 Talin_cent_sf
IPR018979 FERM_N
IPR002558 ILWEQ_dom
IPR037438 Talin1/2-RS
IPR035963 FERM_2
IPR019749 Band_41_domain
IPR032425 FERM_f0
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR000299 FERM_domain
IPR015009 Vinculin-bd_dom
IPR015224 Talin_cent
IPR019748 FERM_central
IPR036723 Alpha-catenin/vinculin-like_sf
IPR030224 Sla2_fam
IPR011993 PH-like_dom_sf
IPR036476 Talin_cent_sf
IPR018979 FERM_N
IPR002558 ILWEQ_dom
IPR037438 Talin1/2-RS
IPR035963 FERM_2
IPR019749 Band_41_domain
IPR032425 FERM_f0
IPR014352 FERM/acyl-CoA-bd_prot_sf
Gene 3D
ProteinModelPortal
PDB
3IVF
E-value=3.93008e-148,
Score=1352
Ontologies
GO
PANTHER
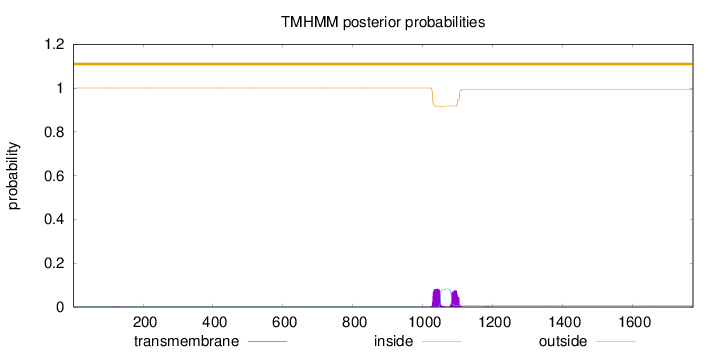
Topology
Length:
1774
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.55189000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00033
outside
1 - 1774
Population Genetic Test Statistics
Pi
273.45636
Theta
180.562406
Tajima's D
1.983299
CLR
0.498672
CSRT
0.880605969701515
Interpretation
Uncertain
