Gene
KWMTBOMO00987
Pre Gene Modal
BGIBMGA001369
Annotation
PREDICTED:_trypsin_delta/gamma-like_[Papilio_polytes]
Full name
Serine protease 3
Alternative Name
Protein Jonah 99Ci
Location in the cell
PlasmaMembrane Reliability : 2.517
Sequence
CDS
ATGTCAGAACAACAAACATCTACAACAAATAACCTAATAAAAATTATTAAAATGAAAGCCGGTTGTATTATACTGACAGTGTTAATAATCATAGCAATCTTGGCAGTTTGTGTTATGGTAAAATTCGCCACTGACATAACAATGGAGGAGAGGAAAAGAGACTCAAACACTGGCTCCGTGTACCAACTCGCGAAAACAATGGGCATCATAAACCTCAAAGAGAACTATTCACTGAACAACGCGGAAAGATTTCCTTACGTTGGCGCAGTAGTGGCTAACACCTCGGGGATTTGGTCCTTTTCATGTTTCGCCAGCGTAATTTTGGTTAAATGGATTGTAACTTCGGCGCATTGCGTGAAACGTGACACGAAACACAGGCTCCTGCTCTTTCACGATTACACGAAGAATTACAGTCACACGTACCCCGTTTTATACTGGAAGCTCCATCAGAAATATAACGTGAGCAAGCCCACGCTCAGACACGACGTGGCCGTTGCAAAATTGAATGTAGACTTCTATCCTTTTTCGACAAAAGCCTCCGTTTTTGACCGGAACCCGCCAGAAACGGACGTCCTTACCGCCGTTCTGTGGAAAACTGTAGCGGCCATAGACAAGAAAATGTATTTGACGAACGATTTTGATAAAATCGAAGTCCAAATCACGAGTTACAACAGATGCTTCGAATCGTATGGCGTAGACTTGGATGCCTCTTTGATTTGCATCGATTTAACGGAGTATGAAGAGTGTTTTGTTCACGAGTTCGGCCCTCTCTACTACGAGGATAAAATCGTGGGCGTGCTGGCTGTGAAGCCGAGGGACTGCGACACCAAATACGCTATCTTTACAAACGTGTCTTTTTATAGAGATTGGATATTGAAATCGACTGGTACAACTTGTTACGGATGA
Protein
MSEQQTSTTNNLIKIIKMKAGCIILTVLIIIAILAVCVMVKFATDITMEERKRDSNTGSVYQLAKTMGIINLKENYSLNNAERFPYVGAVVANTSGIWSFSCFASVILVKWIVTSAHCVKRDTKHRLLLFHDYTKNYSHTYPVLYWKLHQKYNVSKPTLRHDVAVAKLNVDFYPFSTKASVFDRNPPETDVLTAVLWKTVAAIDKKMYLTNDFDKIEVQITSYNRCFESYGVDLDASLICIDLTEYEECFVHEFGPLYYEDKIVGVLAVKPRDCDTKYAIFTNVSFYRDWILKSTGTTCYG
Summary
Description
Its major function may be to aid in digestion.
Similarity
Belongs to the peptidase S1 family.
Keywords
Complete proteome
Disulfide bond
Hydrolase
Protease
Reference proteome
Serine protease
Signal
Zymogen
Feature
chain Serine protease 3
Uniprot
H9IVT8
A0A2A4JPP8
A0A2A4JNU5
A0A2H1VEG8
A0A194RMX5
A0A194PSG1
+ More
A0A212FBU0 A0A2A4JQ60 A0A2H1VEB7 A0A182T985 A0A182YKS8 Q7JYN3 Q9VRS4 A0A1E1WAD4 I2MZM6 A0A1I8PU71 B4HNG4 A0A2C9GRR7 Q7QIZ2 A0A3B5AXE9 A0A182KQG4 A0A369SEQ2 A0A226D5J2 A0A2J7PMZ2 B3LYV0 B4HUM3 G3T0B7 A0A034WE85 P17207 A0A0C9QDL4 T1K0Z0 A0A2D8RD56 B4NLE7 A0A182UM13 A0A0A1WN81 G1TDD9 W8CDM8 G1TI74 B3MRG5 C3YMS0 A0A1I8NCK9 B0WLB8 A0A2P6L5W1 B4QIV9 A0A1I8NX67 A0A0L0CER1 B4QIV6 A0A1I8M6P2 G1SV25 A0A1Q3FSI0 B3RP99 A0A1W0WIL2 A0A034WRC1 W5JVV6 K7JAG2 A0A0L0CLG5 B3NCC1 B3P825 B3NCC0 A0A0P5UZA0 A0A0P5ZLH8
A0A212FBU0 A0A2A4JQ60 A0A2H1VEB7 A0A182T985 A0A182YKS8 Q7JYN3 Q9VRS4 A0A1E1WAD4 I2MZM6 A0A1I8PU71 B4HNG4 A0A2C9GRR7 Q7QIZ2 A0A3B5AXE9 A0A182KQG4 A0A369SEQ2 A0A226D5J2 A0A2J7PMZ2 B3LYV0 B4HUM3 G3T0B7 A0A034WE85 P17207 A0A0C9QDL4 T1K0Z0 A0A2D8RD56 B4NLE7 A0A182UM13 A0A0A1WN81 G1TDD9 W8CDM8 G1TI74 B3MRG5 C3YMS0 A0A1I8NCK9 B0WLB8 A0A2P6L5W1 B4QIV9 A0A1I8NX67 A0A0L0CER1 B4QIV6 A0A1I8M6P2 G1SV25 A0A1Q3FSI0 B3RP99 A0A1W0WIL2 A0A034WRC1 W5JVV6 K7JAG2 A0A0L0CLG5 B3NCC1 B3P825 B3NCC0 A0A0P5UZA0 A0A0P5ZLH8
EC Number
3.4.21.-
Pubmed
19121390
26354079
22118469
25244985
10731132
12537568
+ More
12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22740677 17994087 12364791 14747013 17210077 20966253 30042472 25348373 12537569 2469005 25830018 21993624 24495485 18563158 25315136 22936249 26108605 18719581 20920257 23761445 20075255
12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22740677 17994087 12364791 14747013 17210077 20966253 30042472 25348373 12537569 2469005 25830018 21993624 24495485 18563158 25315136 22936249 26108605 18719581 20920257 23761445 20075255
EMBL
BABH01038071
NWSH01000881
PCG73736.1
PCG73735.1
ODYU01002097
SOQ39176.1
+ More
KQ460152 KPJ17351.1 KQ459593 KPI96376.1 AGBW02009276 OWR51193.1 PCG73734.1 SOQ39175.1 AY075321 AAL68188.2 AE014296 AF512510 AAF50716.1 AAM46864.1 GDQN01007155 JAT83899.1 AJSZ01000583 EIF90223.1 CH480816 EDW47400.1 APCN01000169 AAAB01008807 EAA04161.3 NOWV01000016 RDD45408.1 LNIX01000035 OXA40118.1 NEVH01023960 PNF17686.1 CH902617 EDV44066.1 CH480817 EDW50644.1 GAKP01006497 JAC52455.1 AE014297 AY070605 M24380 GBYB01001399 JAG71166.1 CAEY01001145 NZNF01000029 MAM87894.1 CH964272 EDW84350.1 GBXI01013778 JAD00514.1 AAGW02059878 GAMC01001671 JAC04885.1 CH902622 EDV34370.1 GG666531 EEN58411.1 DS232962 DS231983 EDS27062.1 EDS30402.1 MWRG01001579 PRD33950.1 CM000363 CM002912 EDX09384.1 KMY97838.1 JRES01000501 KNC30741.1 EDX09381.1 GFDL01004672 JAV30373.1 DS985242 EDV27599.1 MTYJ01000095 OQV15055.1 GAKP01002242 JAC56710.1 ADMH02000310 ETN67024.1 JRES01000234 KNC33091.1 CH954178 EDV51079.1 CH954182 EDV53152.1 EDV51078.1 GDIP01123236 JAL80478.1 GDIP01055606 JAM48109.1
KQ460152 KPJ17351.1 KQ459593 KPI96376.1 AGBW02009276 OWR51193.1 PCG73734.1 SOQ39175.1 AY075321 AAL68188.2 AE014296 AF512510 AAF50716.1 AAM46864.1 GDQN01007155 JAT83899.1 AJSZ01000583 EIF90223.1 CH480816 EDW47400.1 APCN01000169 AAAB01008807 EAA04161.3 NOWV01000016 RDD45408.1 LNIX01000035 OXA40118.1 NEVH01023960 PNF17686.1 CH902617 EDV44066.1 CH480817 EDW50644.1 GAKP01006497 JAC52455.1 AE014297 AY070605 M24380 GBYB01001399 JAG71166.1 CAEY01001145 NZNF01000029 MAM87894.1 CH964272 EDW84350.1 GBXI01013778 JAD00514.1 AAGW02059878 GAMC01001671 JAC04885.1 CH902622 EDV34370.1 GG666531 EEN58411.1 DS232962 DS231983 EDS27062.1 EDS30402.1 MWRG01001579 PRD33950.1 CM000363 CM002912 EDX09384.1 KMY97838.1 JRES01000501 KNC30741.1 EDX09381.1 GFDL01004672 JAV30373.1 DS985242 EDV27599.1 MTYJ01000095 OQV15055.1 GAKP01002242 JAC56710.1 ADMH02000310 ETN67024.1 JRES01000234 KNC33091.1 CH954178 EDV51079.1 CH954182 EDV53152.1 EDV51078.1 GDIP01123236 JAL80478.1 GDIP01055606 JAM48109.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000075901
+ More
UP000076408 UP000000803 UP000005940 UP000095300 UP000001292 UP000075840 UP000007062 UP000261400 UP000075882 UP000253843 UP000198287 UP000235965 UP000007801 UP000007646 UP000015104 UP000225829 UP000007798 UP000075903 UP000001811 UP000001554 UP000095301 UP000002320 UP000000304 UP000037069 UP000009022 UP000000673 UP000002358 UP000008711
UP000076408 UP000000803 UP000005940 UP000095300 UP000001292 UP000075840 UP000007062 UP000261400 UP000075882 UP000253843 UP000198287 UP000235965 UP000007801 UP000007646 UP000015104 UP000225829 UP000007798 UP000075903 UP000001811 UP000001554 UP000095301 UP000002320 UP000000304 UP000037069 UP000009022 UP000000673 UP000002358 UP000008711
Interpro
IPR001254
Trypsin_dom
+ More
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR012224 Pept_S1A_FX
IPR018056 Kringle_CS
IPR038178 Kringle_sf
IPR000001 Kringle
IPR013806 Kringle-like
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR007280 Peptidase_C_arc/bac
IPR009003 Peptidase_S1_PA
IPR033116 TRYPSIN_SER
IPR018114 TRYPSIN_HIS
IPR001314 Peptidase_S1A
IPR012224 Pept_S1A_FX
IPR018056 Kringle_CS
IPR038178 Kringle_sf
IPR000001 Kringle
IPR013806 Kringle-like
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR007280 Peptidase_C_arc/bac
Gene 3D
ProteinModelPortal
H9IVT8
A0A2A4JPP8
A0A2A4JNU5
A0A2H1VEG8
A0A194RMX5
A0A194PSG1
+ More
A0A212FBU0 A0A2A4JQ60 A0A2H1VEB7 A0A182T985 A0A182YKS8 Q7JYN3 Q9VRS4 A0A1E1WAD4 I2MZM6 A0A1I8PU71 B4HNG4 A0A2C9GRR7 Q7QIZ2 A0A3B5AXE9 A0A182KQG4 A0A369SEQ2 A0A226D5J2 A0A2J7PMZ2 B3LYV0 B4HUM3 G3T0B7 A0A034WE85 P17207 A0A0C9QDL4 T1K0Z0 A0A2D8RD56 B4NLE7 A0A182UM13 A0A0A1WN81 G1TDD9 W8CDM8 G1TI74 B3MRG5 C3YMS0 A0A1I8NCK9 B0WLB8 A0A2P6L5W1 B4QIV9 A0A1I8NX67 A0A0L0CER1 B4QIV6 A0A1I8M6P2 G1SV25 A0A1Q3FSI0 B3RP99 A0A1W0WIL2 A0A034WRC1 W5JVV6 K7JAG2 A0A0L0CLG5 B3NCC1 B3P825 B3NCC0 A0A0P5UZA0 A0A0P5ZLH8
A0A212FBU0 A0A2A4JQ60 A0A2H1VEB7 A0A182T985 A0A182YKS8 Q7JYN3 Q9VRS4 A0A1E1WAD4 I2MZM6 A0A1I8PU71 B4HNG4 A0A2C9GRR7 Q7QIZ2 A0A3B5AXE9 A0A182KQG4 A0A369SEQ2 A0A226D5J2 A0A2J7PMZ2 B3LYV0 B4HUM3 G3T0B7 A0A034WE85 P17207 A0A0C9QDL4 T1K0Z0 A0A2D8RD56 B4NLE7 A0A182UM13 A0A0A1WN81 G1TDD9 W8CDM8 G1TI74 B3MRG5 C3YMS0 A0A1I8NCK9 B0WLB8 A0A2P6L5W1 B4QIV9 A0A1I8NX67 A0A0L0CER1 B4QIV6 A0A1I8M6P2 G1SV25 A0A1Q3FSI0 B3RP99 A0A1W0WIL2 A0A034WRC1 W5JVV6 K7JAG2 A0A0L0CLG5 B3NCC1 B3P825 B3NCC0 A0A0P5UZA0 A0A0P5ZLH8
PDB
1AZZ
E-value=4.19558e-09,
Score=145
Ontologies
GO
Topology
Subcellular location
Secreted
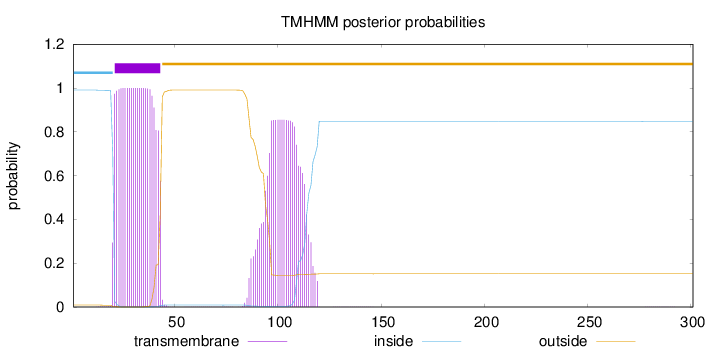
Length:
301
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
41.4838000000002
Exp number, first 60 AAs:
22.37177
Total prob of N-in:
0.99128
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 301
Population Genetic Test Statistics
Pi
318.163849
Theta
192.760694
Tajima's D
1.926612
CLR
0
CSRT
0.866806659667017
Interpretation
Uncertain
