Gene
KWMTBOMO00980
Pre Gene Modal
BGIBMGA001365
Annotation
PREDICTED:_uncharacterized_protein_LOC106107297_isoform_X3_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 3.735
Sequence
CDS
ATGTTTTATTTTCCAGCAAGAAAATGTGCTTATATAGCTAAAATCGGAATATTGTTTACATTAGTGATCGCTTTAATAACTTTCTTCGAACAATGGCGAGGCGGTAAACGGTCCGTGGACGCGACTTTTTCTGATCCGATTGAGGCTGAATTTGAAAGACAAATTTTGGAGGATGAAGCCCGAATTATCCCAGGGTTGGGAGACGGGGGTGTTGCGGCATACCTGACCGGGGAAGACAAGCGTCTTGGCGAAGAGTCTGAGAAGAAACTGGCCATGAATGTGCATCTCAGTGATAGGATAGCGTATAACCGTACTTTGAAAGATTACCGTAACCCAGCTTGCCAGCGAGTGGTCTACGATGCAGAGCTCCCCTCTGTGTCTGTCATCCTGATATTCCACAATGAGCCCTATTCAGTCGTCATAAGAACAATATGGAGTGTTATTAACAGTGCCAGGAGAGATCAACCATGGTATTCGAAAGCTAATTTTGTTGAAAGAGGAACAGGCAGAACTATGCAGTTGGGTTACCCGGGTCAAGACCCTACATCATCATTAGTTTACTTAAAGGAGATAATCCTGGTCGACGATAACTCAACTCTACCAGAGCTTAAAGGAAAGCTCAGTTACTACGTAAAGACCCGTCTGCCTCCTGACCTGATAAGGATTTTAAGATTGCCAGATCGGCAGGGTCTGATGAAGGCCCGGCTGGCGGGGGCGAGGGTCGCGAGAGGTGACGTCCTGGTATTCCTGGACGCTCACTGCGAAACGGGCGCCGATTGGCTGCGGCCCCTCTTGCAGAGGATCACGCACAAGAGAGATTCGGTCCTCACTCCGCTCATAGACGTTGTAGACCAGAGCAGCTTCAAGCTGGAGGCTGCTGAATACTTCCAGGTGGGAGGTTTTACCTTCACTGGTCATTTCACCTGGATTGACGTTCCCGAACGCGAGAAGAAACGTCGCGGCTCAGACATCGCGCCGACCTGGTCCCCAACAATGGCAGGGGGTTTGTTTGCCATCAACCGGGCTTACTTTTGGGAGCTGGGCGCCTACGATGAACAGATGGCTGGCTGGGGCGGTGAGAATCTGGAGATGTCGTTCAGGATATGGCAATGCGGTGGAACACTAGAGACGGTTCCATGCTCACGAGTCGGCCACGTCTTCAGGAGTTTCCATCCCTACGGACTGCCCGCCCAAAGCGACACACACGGTATAAACACGGCTAGGATGGCGGAAGTTTGGATGGATGAATACGCTGAGCTGTTCTACTTGCACCGACCGGACCTCAGGAACAACCCGAAGATAGGCGACGTGACCCATCGGAAGGTGCTGCGCGAGAAACTAAAGTGTAAAGACTTCCAGTGGTACCTGGACAACGTCTACGAGGACAAGTTCGTGCCGGTGCGCGACGTCTACGGCTTCGGGCGATTCAAAAACCCCGCGTCCGGGTTCTGTTTGGACACGATGCAGAGGGACGTAGAGGCGTCGTCTCTGGGCGTCTATCCCTGTCACTCCACACTGCAGGCCACTCAGTATCTGTCTCTTTCACTCCGCGGGGAACTCCGGGATGAGGAGAAATGCGCTCAGCTGCAATATACTAGAAGCGAGAATGATGTGAACGATGACACCGCCAGGAGAGTGCTGATGACCAGCTGCCACGGGAAACAAAGGGGCCAGAAATGGAAATACATTCCGTCCACCAGTCAACTCCAGCACGTCGACTCGGGCTTGTGCCTGGACGCGGGCTTCGAGGTCGGAGCAGACGTCACGGCGAGGGCCTGCAGCGGGAAGGTCCAGCAACGTTGGCTGATCGATTACGCGGAGTACAACCAGTTCAAAGGGAAAAACGGTGAGACGAGTGAGAGAGACGAGAAGCTACGAAGATTGAGGGGACAGAGGAGAAAATCTCGCTCGCTGCTATCTCACGAGGGGGACGCGCAGCAACGCTCCCACGAGCACCGCAGCAGGAAGAGGAAACAGAAGAGAAGGAAGAACAAGAGTAAAAAGAAACGCAAGCGCAAGGACAAGTTCATACTGACGCTGCTGCGGAGCCGCCCCGGGGCGCCCGACGAGCACCTGGAGGCCGAGCTGTACTGCCGCCACGCGCGCCTGCACCCCAACAACAGCTTCGTCAGGGACCTCGTTGCCATCCTCAACGATAAGGATGTCAAGGTGATCAACAACGGGCGTCTGTTCGAGCGCGGCGGGGTCACCGTACTCCGGGACGCCGAAATCAAGAAAATGAAGATCAAGAGTCATGACGACATCGATGACGTCACGACCTCAAAACCTAGCCATAAAAAGTTTAAGACAACGCCATCGGTCAAACGCATCCCCGCCGTGAACGTAGCGACGCTGGCGCCCCCCGCCCACCGCGGCAAGCAACTCAACAAGAAGATTATCATTGAAGAGTTCGTGGCCGTCGCACCGGACGAGAAGACCAAGAAACTGAAGAAGCGTAAAAGAAAACACAGAAGACGCACCACGGTCCCCCCCGCCGCGTACGGAGAGGGGGAGGGAGTGCTCGACCATAACACAGAGAGGAACGCACACGCGTCTGTAGAGCTGACGGACGCGCCACCCTCGTCCGCCGAGGACACCCCCATGCTCATGGGGGCCGAGCGGGGCGCCGACAGGAAGGTCAACGAGCTGCTCAGAGACGACAGCCGTGAGAACTGGGACGTGGGTGAGTCTCGCTTCCGGCGGAACAACTCGCAGACGCTGCTGCAGCCGCTGCGGAGGCTCACCACCGACGAGGGCGGCGCCGACAGCGGCGACGAGTCGTCGAAACAAGATGACGTCACGCCTGAGCGGGATGAGGACAGGAAACCGGTCAAGGTCGTGCTCCGCAGCAACCTGACCTTCAACATCGGCGATGATTTCTTCCCGTGGAGACCGAGGGAAGATGAGAAAGACGACACCGCCTCCACAAAAGATGCGAAGGCACAACAGATACAGCTGATCATAAAGCCTAACCCGGGAATACTCGTAGACGACCAGAGGCCCGCCGCTGACGTCAGGAGTGACGTCACGAAAACGTCAAACAAGAGGATTGACAAACACAAGGACCTGTCAAATTATGACAGGAGTCGCATTAGACATGTGTCAGGCGATGGGAATATAGATCAGCGTCAGGAAGACAGCTTCGAGGATGATAACGTGTTGAGGGGTGGCGGCAAGGCGAGCAGCACTACCGATGATAAGTCCGATGGCTCGTCCAGTTCCTCTGAGAGTTGCGATGATTGA
Protein
MFYFPARKCAYIAKIGILFTLVIALITFFEQWRGGKRSVDATFSDPIEAEFERQILEDEARIIPGLGDGGVAAYLTGEDKRLGEESEKKLAMNVHLSDRIAYNRTLKDYRNPACQRVVYDAELPSVSVILIFHNEPYSVVIRTIWSVINSARRDQPWYSKANFVERGTGRTMQLGYPGQDPTSSLVYLKEIILVDDNSTLPELKGKLSYYVKTRLPPDLIRILRLPDRQGLMKARLAGARVARGDVLVFLDAHCETGADWLRPLLQRITHKRDSVLTPLIDVVDQSSFKLEAAEYFQVGGFTFTGHFTWIDVPEREKKRRGSDIAPTWSPTMAGGLFAINRAYFWELGAYDEQMAGWGGENLEMSFRIWQCGGTLETVPCSRVGHVFRSFHPYGLPAQSDTHGINTARMAEVWMDEYAELFYLHRPDLRNNPKIGDVTHRKVLREKLKCKDFQWYLDNVYEDKFVPVRDVYGFGRFKNPASGFCLDTMQRDVEASSLGVYPCHSTLQATQYLSLSLRGELRDEEKCAQLQYTRSENDVNDDTARRVLMTSCHGKQRGQKWKYIPSTSQLQHVDSGLCLDAGFEVGADVTARACSGKVQQRWLIDYAEYNQFKGKNGETSERDEKLRRLRGQRRKSRSLLSHEGDAQQRSHEHRSRKRKQKRRKNKSKKKRKRKDKFILTLLRSRPGAPDEHLEAELYCRHARLHPNNSFVRDLVAILNDKDVKVINNGRLFERGGVTVLRDAEIKKMKIKSHDDIDDVTTSKPSHKKFKTTPSVKRIPAVNVATLAPPAHRGKQLNKKIIIEEFVAVAPDEKTKKLKKRKRKHRRRTTVPPAAYGEGEGVLDHNTERNAHASVELTDAPPSSAEDTPMLMGAERGADRKVNELLRDDSRENWDVGESRFRRNNSQTLLQPLRRLTTDEGGADSGDESSKQDDVTPERDEDRKPVKVVLRSNLTFNIGDDFFPWRPREDEKDDTASTKDAKAQQIQLIIKPNPGILVDDQRPAADVRSDVTKTSNKRIDKHKDLSNYDRSRIRHVSGDGNIDQRQEDSFEDDNVLRGGGKASSTTDDKSDGSSSSSESCDD
Summary
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
1XHB
E-value=3.39483e-101,
Score=945
Ontologies
PATHWAY
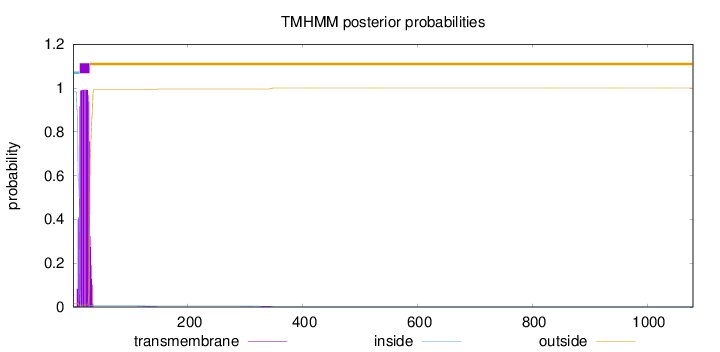
Topology
Length:
1080
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.11641
Exp number, first 60 AAs:
19.95649
Total prob of N-in:
0.98374
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 29
outside
30 - 1080
Population Genetic Test Statistics
Pi
327.253031
Theta
183.741591
Tajima's D
2.66819
CLR
0.053462
CSRT
0.95685215739213
Interpretation
Uncertain
