Pre Gene Modal
BGIBMGA001364
Annotation
PREDICTED:_vacuolar_protein_sorting-associated_protein_53_homolog_[Papilio_xuthus]
Full name
Vacuolar protein sorting-associated protein 53 homolog
Location in the cell
Nuclear Reliability : 2.894
Sequence
CDS
ATGGATTCGAATGATGCTACGGATGATAAAGACAGCGGGCCAGAAGTTCAAATTAACTTTCCGAGTTCCGTAATGAGCCGCATAGAGGAATTAATGGGAGGAACGGAACAGTTCGATAGCGATGACTTTGATGCAGTGGCGTACATCAACAGAGTGTTCCCAACTGAGCAATCTCTTTCTGGAGTGGAAAGTGCTGTGGCTCGCTGTGAATTTAGACTCTCAGGAGTGGAGCAAGATATTAGAAGACTGGTCCGAGCACAGGCTGAACAGAGGGAAGCTGGACAGGAGGCACTAACGGAAGCTCAGAGATGTATAGCAGAGTTAGCTTTGCAGGTCGCGGATATAAACAAGAAAGCTGAAAGAAGCGAAAGCATGGTCAGGGAGATAACTTGCGAGATAAAGCAGTTGGACTGCGCTAAGTGTAATCTGACCGCTGCTATTACCGCCCTCAACCATCTGCACATGCTGGCTGGCGGTGTGGACACCCTCACAACAATGACGAAGAACCGCCAGTACAAGGAAATAGCGTTGCCCATGCAGGCTATTATGGAGGTCTTACAGCACTTTGAATGTTACCGAGACATCCGCGAACTCAGCGTCTTAAGGGATCAAGTGCACTCGATAAGGCACGATTTGGGCAATCAAATATTAGCTGATTTTAAACAAGCCTTTACAGGCGGCAGTAAAAGCAACATAACTCATAGGACGTTATCGGAGGCTTGCGGCGTCGTCGACATCTTAGACCCGAAGGTCAAACAGGATCTGTTAAAGTGGTTCATCAGCATGCAACTACAGGAATACCAGCACCTGTACTCGTCGGAGCAGGAGTGCGCCTGGCTCACTCACATCGAGCGACGGTACGCGTGGCTCAAGAAGCATCTCCTCACGTTCGAAGACACCCTCGGAAATATATTCCCTTACCAGTGGGCTCTGAGCGAGAACATAGCCAGGCAGTTCTGTAAGATCACCTCAACGGAATTAAGCAATCTAATGGCGGCGCGGAGAAACGAATTAGACGTGAAACTGCTGCTGTACGCGATACAGAACACTTATAACTTCGAATTACTGCTGCACAAGAGGTTCACAGGTACAGACCTGAAGCCTGACAATGTAGACACGAACTCTAAACAGCCGTTCGATAATGAAACGAAAGATAACACCGCAGACTCGGACGGCCAGCAGGCGGAAGGCTCGTCCAGCAGTTGGGTCGGGCTGATAGGGTCCTGCTTCGAGCCCTATCTCTCATTGTACATCAACAGTCTGGACTCGAACCTGAGCTCGCTCATCGAGAGATTCGTGCAGGATGCAAAGACGTCGCCCTCTGAAGCCAACATGACCGCCAGTGGTGAAGGGGCCGTCATATCCAGCTGCGCGGATTTGTTCCTCTTCTACAAGAAATGCTTGGTGCAATGCGCCAGGCTCACCACAGGGGAACCGATGCTGGAACTAGCGGGAGTATTCCAGAAGTATCTACGCGAGTACGCCAGCTCTGTGCTGCAAGCTGCCCTGCCCAGGACCAACCCGACGTCCCTCCTCTCGCTGTCGCTAAGTGGCAACACCGTGCCGAGCATAGTCACGAACTTACACACGCTACTCAGAGATGAAGCCGGTAACGTTTCCAGATACACAAAACAGGAAATCTCAAAGATAACAAGCGTGATCACGACCTCGGAGTACTGCCTCGAGACCACGATACATCTGGAGCAGAAACTCAAGGAGAAGGTCAGTCCGAAGCTGTCCGACAAAATCGATTTGACACCGGAACAGGATTTGTTCCACAAGATCATCAGCAACTGCATCCAGATGCTGGTCCAGGACCTGGAGACGGCCTGCGAGCCGGCCCTCCAGAGCATGACCAAGATATCCTGGCTGAGCTTCGACAACGTAGGCGACCAGAGTTCTTACGTGACTCAGATAATAATGCACTTGAAGAACACCGTGCCGAACCTGAGGGACAACTTATCGTCGTCCAGGAAGTACTTCACGCAGTTCTGCATACGATTCGCTAATTCTTTCATACCGAAGTTCATTCAGAACGTGTACAAGTGCAAGCCCATCTCCACGGTCGGCTCTGAGCAGCTGTTGCTTGACACGCACATGCTGAAGACGGCGCTGTTAGAACTACCCTCAATCGGGTCGGAGGTCAAAAGGGCCGCACCCGCAACCTACACGAAAGTCGTCCTCAAGCTGATGACGAAAGCGGAAATGGTTCTCAAATTGGTGATGGCCCCCCTCGAGGGGAATCTGGAGGGGTTCGTCACGCAGTTCGTCAAATTGCTACCGGATTGCTCCTTGGCTGAGTTCCACAAGGTTTTAGACATGAAAGGCGCTAAGCTATCTAAAACTCAGATGAGCAACTTGGATGCTGTCTTCAAGAATGCCTGCAAGTCTGCCGATGTTAAATAG
Protein
MDSNDATDDKDSGPEVQINFPSSVMSRIEELMGGTEQFDSDDFDAVAYINRVFPTEQSLSGVESAVARCEFRLSGVEQDIRRLVRAQAEQREAGQEALTEAQRCIAELALQVADINKKAERSESMVREITCEIKQLDCAKCNLTAAITALNHLHMLAGGVDTLTTMTKNRQYKEIALPMQAIMEVLQHFECYRDIRELSVLRDQVHSIRHDLGNQILADFKQAFTGGSKSNITHRTLSEACGVVDILDPKVKQDLLKWFISMQLQEYQHLYSSEQECAWLTHIERRYAWLKKHLLTFEDTLGNIFPYQWALSENIARQFCKITSTELSNLMAARRNELDVKLLLYAIQNTYNFELLLHKRFTGTDLKPDNVDTNSKQPFDNETKDNTADSDGQQAEGSSSSWVGLIGSCFEPYLSLYINSLDSNLSSLIERFVQDAKTSPSEANMTASGEGAVISSCADLFLFYKKCLVQCARLTTGEPMLELAGVFQKYLREYASSVLQAALPRTNPTSLLSLSLSGNTVPSIVTNLHTLLRDEAGNVSRYTKQEISKITSVITTSEYCLETTIHLEQKLKEKVSPKLSDKIDLTPEQDLFHKIISNCIQMLVQDLETACEPALQSMTKISWLSFDNVGDQSSYVTQIIMHLKNTVPNLRDNLSSSRKYFTQFCIRFANSFIPKFIQNVYKCKPISTVGSEQLLLDTHMLKTALLELPSIGSEVKRAAPATYTKVVLKLMTKAEMVLKLVMAPLEGNLEGFVTQFVKLLPDCSLAEFHKVLDMKGAKLSKTQMSNLDAVFKNACKSADVK
Summary
Description
Acts as component of the GARP complex that is involved in retrograde transport from early and late endosomes to the trans-Golgi network (TGN). Acts as component of the EARP complex that is involved in endocytic recycling.
Cofactor
Mn(2+)
Subunit
Component of the Golgi-associated retrograde protein (GARP) complex. Component of the endosome-associated retrograde protein (EARP) complex.
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Belongs to the VPS53 family.
Belongs to the VPS53 family.
Keywords
Coiled coil
Complete proteome
Endosome
Golgi apparatus
Membrane
Protein transport
Reference proteome
Transport
Feature
chain Vacuolar protein sorting-associated protein 53 homolog
Uniprot
H9IVT3
A0A194PTR3
A0A194RHQ9
A0A212FDA0
A0A2W1BFT3
D2A3P8
+ More
A0A067RBC2 A0A2J7PF47 A0A1W4XMU3 A0A1Y1L9F9 T1IEK0 N6URT6 A0A0N0U3P0 A0A0L7RFC1 A0A088A2I1 A0A0A9XAK6 A0A224XIW5 A0A310S573 A0A026X1V8 A0A069DWG6 A0A2R7VYU7 A0A0V0G9Z7 C3Y0K3 A0A154PBP9 A0A232FGE0 K7J7Y8 E2BNX9 A0A1B6E0N4 T1J5E2 A0A195E2P9 F4WYI8 A0A151I2P3 A0A158P1G8 A0A087T6F5 A0A195F4G9 A0A195CSI7 A0A1S3DUQ1 A0A2R5LJE5 A0A293MHW9 K1Q8K0 A0A2T7PHL6 V5HWX5 A0A1B6IVP8 A0A210Q0M7 A0A1L8DW08 V4AJT6 W5KN75 A0A224YSH5 A0A0K8TM60 A0A1L8HG35 A0A3B4DJP8 A0A2R2MS88 A0A218UUP7 F6P6T6 Q5ZLD7 A0A226NXN2 E7FAG0 A0A0P5PA70 A0A1L8H870 A0A164QN31 A0A1V4K267 A0A2G8LEP1 A0A0P5XBY7 A0A2S2QG09 W5MAP0 A0A3P9IQV4 V9KCP0 A0A0L0CG23 A0A087R014 A0A087VBJ9 A0A091UMF0 A0A091T6J2 A0A093N8A2 A0A336KA85 A0A093FJA0 A0A091HX73 A0A3P9LFR0 A0A1B0DEF8 E9G261 A0A091UDH5 A0A091M7V2 G3P0E5 A0A1W4YID7 A0A091P9A8 A0A091L0K2 A0A151NAC3 A0A2D0SUY7 A0A091EJD6 F6WN34 R0K9M1 A0A093JN66 A0A093JHW5 M7CAR2 H3APE0 A0A2K5D0V0 U3JT97 A0A091KGI8 A0A1A7XDJ7 L5LLT4 A0A091H6I2
A0A067RBC2 A0A2J7PF47 A0A1W4XMU3 A0A1Y1L9F9 T1IEK0 N6URT6 A0A0N0U3P0 A0A0L7RFC1 A0A088A2I1 A0A0A9XAK6 A0A224XIW5 A0A310S573 A0A026X1V8 A0A069DWG6 A0A2R7VYU7 A0A0V0G9Z7 C3Y0K3 A0A154PBP9 A0A232FGE0 K7J7Y8 E2BNX9 A0A1B6E0N4 T1J5E2 A0A195E2P9 F4WYI8 A0A151I2P3 A0A158P1G8 A0A087T6F5 A0A195F4G9 A0A195CSI7 A0A1S3DUQ1 A0A2R5LJE5 A0A293MHW9 K1Q8K0 A0A2T7PHL6 V5HWX5 A0A1B6IVP8 A0A210Q0M7 A0A1L8DW08 V4AJT6 W5KN75 A0A224YSH5 A0A0K8TM60 A0A1L8HG35 A0A3B4DJP8 A0A2R2MS88 A0A218UUP7 F6P6T6 Q5ZLD7 A0A226NXN2 E7FAG0 A0A0P5PA70 A0A1L8H870 A0A164QN31 A0A1V4K267 A0A2G8LEP1 A0A0P5XBY7 A0A2S2QG09 W5MAP0 A0A3P9IQV4 V9KCP0 A0A0L0CG23 A0A087R014 A0A087VBJ9 A0A091UMF0 A0A091T6J2 A0A093N8A2 A0A336KA85 A0A093FJA0 A0A091HX73 A0A3P9LFR0 A0A1B0DEF8 E9G261 A0A091UDH5 A0A091M7V2 G3P0E5 A0A1W4YID7 A0A091P9A8 A0A091L0K2 A0A151NAC3 A0A2D0SUY7 A0A091EJD6 F6WN34 R0K9M1 A0A093JN66 A0A093JHW5 M7CAR2 H3APE0 A0A2K5D0V0 U3JT97 A0A091KGI8 A0A1A7XDJ7 L5LLT4 A0A091H6I2
Pubmed
19121390
26354079
22118469
28756777
18362917
19820115
+ More
24845553 28004739 23537049 25401762 26823975 24508170 26334808 18563158 28648823 20075255 20798317 21719571 21347285 22992520 25765539 28812685 23254933 25329095 28797301 26369729 27762356 23594743 15642098 24621616 29023486 17554307 24402279 26108605 21292972 22293439 17495919 23624526 9215903
24845553 28004739 23537049 25401762 26823975 24508170 26334808 18563158 28648823 20075255 20798317 21719571 21347285 22992520 25765539 28812685 23254933 25329095 28797301 26369729 27762356 23594743 15642098 24621616 29023486 17554307 24402279 26108605 21292972 22293439 17495919 23624526 9215903
EMBL
BABH01038060
KQ459593
KPI96363.1
KQ460152
KPJ17363.1
AGBW02009096
+ More
OWR51697.1 KZ150094 PZC73648.1 KQ971348 EFA04902.1 KK852571 KDR21126.1 NEVH01026085 PNF14951.1 GEZM01061827 JAV70302.1 ACPB03014069 APGK01019131 KB740092 ENN81447.1 KQ435881 KOX69808.1 KQ414601 KOC69677.1 GBHO01026943 GBRD01003331 GDHC01018923 JAG16661.1 JAG62490.1 JAP99705.1 GFTR01008036 JAW08390.1 KQ771508 OAD52537.1 KK107024 EZA62262.1 GBGD01000521 JAC88368.1 KK854169 PTY12509.1 GECL01001249 JAP04875.1 GG666478 EEN66276.1 KQ434869 KZC09263.1 NNAY01000231 OXU29836.1 GL449511 EFN82571.1 GEDC01014517 GEDC01005827 JAS22781.1 JAS31471.1 JH431859 KQ979709 KYN19440.1 GL888450 EGI60720.1 KQ976538 KYM81313.1 ADTU01006541 KK113646 KFM60694.1 KQ981852 KYN34979.1 KQ977305 KYN03673.1 GGLE01005544 MBY09670.1 GFWV01020569 MAA45297.1 JH818600 EKC30298.1 PZQS01000004 PVD32916.1 GANP01005495 JAB78973.1 GECU01016692 JAS91014.1 NEDP02005302 OWF42265.1 GFDF01003464 JAV10620.1 KB201305 ESO97342.1 GFPF01009411 MAA20557.1 GDAI01002159 JAI15444.1 CM004468 OCT95053.1 MUZQ01000126 OWK57356.1 BX908796 AJ719797 AWGT02000338 OXB71889.1 GDIQ01131476 JAL20250.1 CM004469 OCT92293.1 LRGB01002384 KZS07903.1 LSYS01005108 OPJ78526.1 MRZV01000103 PIK58739.1 GDIP01074339 JAM29376.1 GGMS01007418 MBY76621.1 AHAT01005740 JW863462 AFO95979.1 JRES01000438 KNC31196.1 KL226004 KFM06818.1 KL487157 KFO09991.1 KL409726 KFQ91547.1 KK440511 KFQ70049.1 KL224565 KFW61233.1 UFQS01000107 UFQT01000107 SSW99683.1 SSX20063.1 KK628337 KFV54369.1 KL217847 KFO99742.1 AJVK01032716 AJVK01032717 AJVK01032718 GL732530 EFX86362.1 KK426351 KFQ87860.1 KK524251 KFP67556.1 KK654874 KFQ04190.1 KK549994 KFP33494.1 AKHW03003672 KYO33760.1 KK718508 KFO57181.1 KB742569 EOB06971.1 KL206227 KFV80179.1 KK609846 KFW11467.1 KB521250 EMP37747.1 AFYH01008281 AFYH01008282 AFYH01008283 AFYH01008284 AFYH01008285 AFYH01008286 AFYH01008287 AFYH01008288 AFYH01008289 AFYH01008290 AGTO01000875 KK744034 KFP39669.1 HADW01014530 HADX01007053 SBP15930.1 KB110620 ELK26971.1 KL526823 KFO91451.1
OWR51697.1 KZ150094 PZC73648.1 KQ971348 EFA04902.1 KK852571 KDR21126.1 NEVH01026085 PNF14951.1 GEZM01061827 JAV70302.1 ACPB03014069 APGK01019131 KB740092 ENN81447.1 KQ435881 KOX69808.1 KQ414601 KOC69677.1 GBHO01026943 GBRD01003331 GDHC01018923 JAG16661.1 JAG62490.1 JAP99705.1 GFTR01008036 JAW08390.1 KQ771508 OAD52537.1 KK107024 EZA62262.1 GBGD01000521 JAC88368.1 KK854169 PTY12509.1 GECL01001249 JAP04875.1 GG666478 EEN66276.1 KQ434869 KZC09263.1 NNAY01000231 OXU29836.1 GL449511 EFN82571.1 GEDC01014517 GEDC01005827 JAS22781.1 JAS31471.1 JH431859 KQ979709 KYN19440.1 GL888450 EGI60720.1 KQ976538 KYM81313.1 ADTU01006541 KK113646 KFM60694.1 KQ981852 KYN34979.1 KQ977305 KYN03673.1 GGLE01005544 MBY09670.1 GFWV01020569 MAA45297.1 JH818600 EKC30298.1 PZQS01000004 PVD32916.1 GANP01005495 JAB78973.1 GECU01016692 JAS91014.1 NEDP02005302 OWF42265.1 GFDF01003464 JAV10620.1 KB201305 ESO97342.1 GFPF01009411 MAA20557.1 GDAI01002159 JAI15444.1 CM004468 OCT95053.1 MUZQ01000126 OWK57356.1 BX908796 AJ719797 AWGT02000338 OXB71889.1 GDIQ01131476 JAL20250.1 CM004469 OCT92293.1 LRGB01002384 KZS07903.1 LSYS01005108 OPJ78526.1 MRZV01000103 PIK58739.1 GDIP01074339 JAM29376.1 GGMS01007418 MBY76621.1 AHAT01005740 JW863462 AFO95979.1 JRES01000438 KNC31196.1 KL226004 KFM06818.1 KL487157 KFO09991.1 KL409726 KFQ91547.1 KK440511 KFQ70049.1 KL224565 KFW61233.1 UFQS01000107 UFQT01000107 SSW99683.1 SSX20063.1 KK628337 KFV54369.1 KL217847 KFO99742.1 AJVK01032716 AJVK01032717 AJVK01032718 GL732530 EFX86362.1 KK426351 KFQ87860.1 KK524251 KFP67556.1 KK654874 KFQ04190.1 KK549994 KFP33494.1 AKHW03003672 KYO33760.1 KK718508 KFO57181.1 KB742569 EOB06971.1 KL206227 KFV80179.1 KK609846 KFW11467.1 KB521250 EMP37747.1 AFYH01008281 AFYH01008282 AFYH01008283 AFYH01008284 AFYH01008285 AFYH01008286 AFYH01008287 AFYH01008288 AFYH01008289 AFYH01008290 AGTO01000875 KK744034 KFP39669.1 HADW01014530 HADX01007053 SBP15930.1 KB110620 ELK26971.1 KL526823 KFO91451.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000007266
UP000027135
+ More
UP000235965 UP000192223 UP000015103 UP000019118 UP000053105 UP000053825 UP000005203 UP000053097 UP000001554 UP000076502 UP000215335 UP000002358 UP000008237 UP000078492 UP000007755 UP000078540 UP000005205 UP000054359 UP000078541 UP000078542 UP000079169 UP000005408 UP000245119 UP000242188 UP000030746 UP000018467 UP000186698 UP000261440 UP000085678 UP000197619 UP000000437 UP000000539 UP000198419 UP000076858 UP000190648 UP000230750 UP000018468 UP000265200 UP000037069 UP000053286 UP000053283 UP000054081 UP000054308 UP000265180 UP000092462 UP000000305 UP000007635 UP000192224 UP000050525 UP000221080 UP000052976 UP000002280 UP000053584 UP000031443 UP000008672 UP000233020 UP000016665
UP000235965 UP000192223 UP000015103 UP000019118 UP000053105 UP000053825 UP000005203 UP000053097 UP000001554 UP000076502 UP000215335 UP000002358 UP000008237 UP000078492 UP000007755 UP000078540 UP000005205 UP000054359 UP000078541 UP000078542 UP000079169 UP000005408 UP000245119 UP000242188 UP000030746 UP000018467 UP000186698 UP000261440 UP000085678 UP000197619 UP000000437 UP000000539 UP000198419 UP000076858 UP000190648 UP000230750 UP000018468 UP000265200 UP000037069 UP000053286 UP000053283 UP000054081 UP000054308 UP000265180 UP000092462 UP000000305 UP000007635 UP000192224 UP000050525 UP000221080 UP000052976 UP000002280 UP000053584 UP000031443 UP000008672 UP000233020 UP000016665
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IVT3
A0A194PTR3
A0A194RHQ9
A0A212FDA0
A0A2W1BFT3
D2A3P8
+ More
A0A067RBC2 A0A2J7PF47 A0A1W4XMU3 A0A1Y1L9F9 T1IEK0 N6URT6 A0A0N0U3P0 A0A0L7RFC1 A0A088A2I1 A0A0A9XAK6 A0A224XIW5 A0A310S573 A0A026X1V8 A0A069DWG6 A0A2R7VYU7 A0A0V0G9Z7 C3Y0K3 A0A154PBP9 A0A232FGE0 K7J7Y8 E2BNX9 A0A1B6E0N4 T1J5E2 A0A195E2P9 F4WYI8 A0A151I2P3 A0A158P1G8 A0A087T6F5 A0A195F4G9 A0A195CSI7 A0A1S3DUQ1 A0A2R5LJE5 A0A293MHW9 K1Q8K0 A0A2T7PHL6 V5HWX5 A0A1B6IVP8 A0A210Q0M7 A0A1L8DW08 V4AJT6 W5KN75 A0A224YSH5 A0A0K8TM60 A0A1L8HG35 A0A3B4DJP8 A0A2R2MS88 A0A218UUP7 F6P6T6 Q5ZLD7 A0A226NXN2 E7FAG0 A0A0P5PA70 A0A1L8H870 A0A164QN31 A0A1V4K267 A0A2G8LEP1 A0A0P5XBY7 A0A2S2QG09 W5MAP0 A0A3P9IQV4 V9KCP0 A0A0L0CG23 A0A087R014 A0A087VBJ9 A0A091UMF0 A0A091T6J2 A0A093N8A2 A0A336KA85 A0A093FJA0 A0A091HX73 A0A3P9LFR0 A0A1B0DEF8 E9G261 A0A091UDH5 A0A091M7V2 G3P0E5 A0A1W4YID7 A0A091P9A8 A0A091L0K2 A0A151NAC3 A0A2D0SUY7 A0A091EJD6 F6WN34 R0K9M1 A0A093JN66 A0A093JHW5 M7CAR2 H3APE0 A0A2K5D0V0 U3JT97 A0A091KGI8 A0A1A7XDJ7 L5LLT4 A0A091H6I2
A0A067RBC2 A0A2J7PF47 A0A1W4XMU3 A0A1Y1L9F9 T1IEK0 N6URT6 A0A0N0U3P0 A0A0L7RFC1 A0A088A2I1 A0A0A9XAK6 A0A224XIW5 A0A310S573 A0A026X1V8 A0A069DWG6 A0A2R7VYU7 A0A0V0G9Z7 C3Y0K3 A0A154PBP9 A0A232FGE0 K7J7Y8 E2BNX9 A0A1B6E0N4 T1J5E2 A0A195E2P9 F4WYI8 A0A151I2P3 A0A158P1G8 A0A087T6F5 A0A195F4G9 A0A195CSI7 A0A1S3DUQ1 A0A2R5LJE5 A0A293MHW9 K1Q8K0 A0A2T7PHL6 V5HWX5 A0A1B6IVP8 A0A210Q0M7 A0A1L8DW08 V4AJT6 W5KN75 A0A224YSH5 A0A0K8TM60 A0A1L8HG35 A0A3B4DJP8 A0A2R2MS88 A0A218UUP7 F6P6T6 Q5ZLD7 A0A226NXN2 E7FAG0 A0A0P5PA70 A0A1L8H870 A0A164QN31 A0A1V4K267 A0A2G8LEP1 A0A0P5XBY7 A0A2S2QG09 W5MAP0 A0A3P9IQV4 V9KCP0 A0A0L0CG23 A0A087R014 A0A087VBJ9 A0A091UMF0 A0A091T6J2 A0A093N8A2 A0A336KA85 A0A093FJA0 A0A091HX73 A0A3P9LFR0 A0A1B0DEF8 E9G261 A0A091UDH5 A0A091M7V2 G3P0E5 A0A1W4YID7 A0A091P9A8 A0A091L0K2 A0A151NAC3 A0A2D0SUY7 A0A091EJD6 F6WN34 R0K9M1 A0A093JN66 A0A093JHW5 M7CAR2 H3APE0 A0A2K5D0V0 U3JT97 A0A091KGI8 A0A1A7XDJ7 L5LLT4 A0A091H6I2
Ontologies
GO
PANTHER
Topology
Subcellular location
Golgi apparatus
Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
trans-Golgi network membrane Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
Endosome membrane Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
Recycling endosome Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
trans-Golgi network membrane Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
Endosome membrane Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
Recycling endosome Localizes to the trans-Golgi network as part of the GARP complex, while it localizes to recycling endosomes as part of the EARP complex. With evidence from 1 publications.
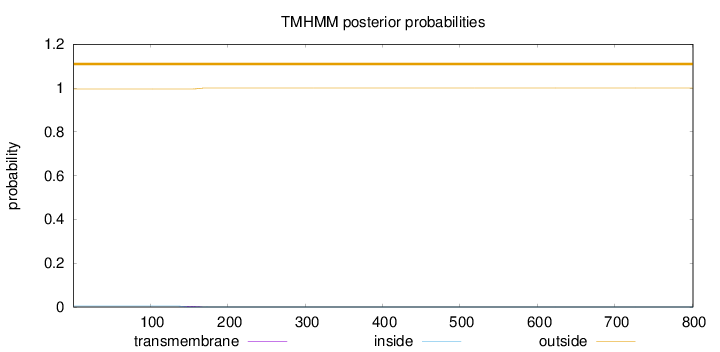
Length:
801
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0835800000000004
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00407
outside
1 - 801
Population Genetic Test Statistics
Pi
197.56881
Theta
166.418846
Tajima's D
1.222821
CLR
0.024094
CSRT
0.71776411179441
Interpretation
Uncertain
