Gene
KWMTBOMO00976
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.992
Sequence
CDS
ATGGAGAAAACCGAAAATACAATTGAAATCCCTATTGAGTGCAAGGCTCCAGCGGAGGAACAGAGAGAAGGAGACGAAAGTAGAACTACAGAAAGTACAGACACTGAATGTGATGATAATGATATCACATATATTCCAGAAGCTTCTTCAGTAAGTGACTCATATGACAGTTTTTCTGAAACAGCATCAGAACCAGAACCTTCATTATTAGAATTGAATCAGGATATTCCAAACAAGCGAGTGAGAAAAATACCTGATAGATTTAGCTATAGTCACTTATGTATTGCAGAATCAGATGATCAAAATATAATGGATATTTCACTCAAGGAAGCTTTAAATGGACCTGAAAGTAAATTTTGGAGATCAAGCATGGAGGAGGAATTAGAATCATTTAAGAAAAATGATGCATGGGAACTTGTGGATAAGCCTCGTGACTCTACAGTAGTGCAGTGCAAATGGGTATTTAAAAAGAAGTATGATAGTGTCGGTAATGTGCGTTATCGTGCTAGATTAGTGGCAAAAGGCTTTTCACAGAAAGCTGGCATAGACTATCATGAAACTTTTTCACCTGTGCTACGTTATACTACTTTAAGATTGTTATTTTCTATAGCAGTAAAACTAGGTTTAGATATACGTCATCTTGATGTCACTACAGCCTTTCTAAATGGTTTTTTGAATGAATCTGTGTATATGCAAAAACCTGTCCTTTATAGAGATTGTAATGATAATGATAACAAAGTATTAAAGCTTAAACGTGCCATATATGGACTTAAGCAATCATCTCGTGCTTGGTATCAGAGAGTTAATGATTACTTAGAGTCTTTAGGATATTTAAAATCTAATTAA
Protein
MEKTENTIEIPIECKAPAEEQREGDESRTTESTDTECDDNDITYIPEASSVSDSYDSFSETASEPEPSLLELNQDIPNKRVRKIPDRFSYSHLCIAESDDQNIMDISLKEALNGPESKFWRSSMEEELESFKKNDAWELVDKPRDSTVVQCKWVFKKKYDSVGNVRYRARLVAKGFSQKAGIDYHETFSPVLRYTTLRLLFSIAVKLGLDIRHLDVTTAFLNGFLNESVYMQKPVLYRDCNDNDNKVLKLKRAIYGLKQSSRAWYQRVNDYLESLGYLKSN
Summary
Uniprot
A0A212F5G1
A0A0A9XV68
A0A0A9XY84
A0A146LH62
W4VRP9
W4VRP2
+ More
A0A0N1ICY1 A0A2N9I4J9 A0A0A9WCG6 A0A2N9ICF5 A0A2N9H9J3 A0A117NI32 A0A2K3PDJ4 A0A2N9FS65 A0A2N9GKN5 A0A2N9HL59 A0A2N9FZX9 A0A2N9HDC0 A0A2N9GJW7 A0A2N9GR48 A0A2N9ITI6 A0A2N9F0H9 A0A2N9HXB8 A0A2N9IMU9 A0A2N9HH69 A0A2N9FGT3 A0A2N9G4F5 A0A2N9FB28 A0A2N9EVG8 A0A2N9FG19 A0A151SR30 A0A2N9GS00 A0A2N9GG65 A0A2N9J3H7 A0A2N9EY13 A0A2N9G1K0 A0A2N9F135 A0A2N9G9I0 A0A1J3IJ96 A0A2N9FUC8 A0A1Y1IGG8 A0A251UJK4 A0A250X157 A0A2K3KEZ8 A0A1Y1IS38
A0A0N1ICY1 A0A2N9I4J9 A0A0A9WCG6 A0A2N9ICF5 A0A2N9H9J3 A0A117NI32 A0A2K3PDJ4 A0A2N9FS65 A0A2N9GKN5 A0A2N9HL59 A0A2N9FZX9 A0A2N9HDC0 A0A2N9GJW7 A0A2N9GR48 A0A2N9ITI6 A0A2N9F0H9 A0A2N9HXB8 A0A2N9IMU9 A0A2N9HH69 A0A2N9FGT3 A0A2N9G4F5 A0A2N9FB28 A0A2N9EVG8 A0A2N9FG19 A0A151SR30 A0A2N9GS00 A0A2N9GG65 A0A2N9J3H7 A0A2N9EY13 A0A2N9G1K0 A0A2N9F135 A0A2N9G9I0 A0A1J3IJ96 A0A2N9FUC8 A0A1Y1IGG8 A0A251UJK4 A0A250X157 A0A2K3KEZ8 A0A1Y1IS38
Pubmed
EMBL
AGBW02010203
OWR48976.1
GBHO01019780
JAG23824.1
GBHO01019781
JAG23823.1
+ More
GDHC01011760 JAQ06869.1 GANO01000659 JAB59212.1 GANO01000660 JAB59211.1 KQ460882 KPJ11482.1 OIVN01004680 SPD18741.1 GBHO01038523 JAG05081.1 OIVN01005285 SPD21760.1 OIVN01003414 SPD11066.1 LKAM01000003 KUM49280.1 ASHM01006021 PNY13361.1 OIVN01001114 SPC90048.1 OIVN01002365 SPD03017.1 OIVN01003620 SPD12483.1 OIVN01001336 SPC92838.1 OIVN01003223 SPD09609.1 OIVN01002001 SPC99720.1 OIVN01002225 SPD01801.1 OIVN01006243 SPD28737.1 OIVN01000459 SPC80612.1 OIVN01004260 SPD16354.1 OIVN01006118 SPD25470.1 OIVN01003416 SPD11073.1 OIVN01000817 SPC86004.1 OIVN01001779 OIVN01003965 SPC97547.1 SPD14614.1 OIVN01000702 SPC84305.1 OIVN01000354 SPC78868.1 OIVN01000822 SPC86055.1 CM003613 KYP57192.1 OIVN01002280 SPD02253.1 OIVN01001872 SPC98448.1 OIVN01006344 SPD31070.1 OIVN01000403 SPC79640.1 OIVN01001384 SPC93353.1 OIVN01000452 SPC80504.1 OIVN01001628 SPC96029.1 GEVM01025909 JAU80029.1 OIVN01001171 SPC90720.1 DF237331 GAQ87846.1 CM007895 OTG23274.1 BEGY01000018 GAX76480.1 ASHM01093982 PNX64819.1 DF238519 GAQ93484.1
GDHC01011760 JAQ06869.1 GANO01000659 JAB59212.1 GANO01000660 JAB59211.1 KQ460882 KPJ11482.1 OIVN01004680 SPD18741.1 GBHO01038523 JAG05081.1 OIVN01005285 SPD21760.1 OIVN01003414 SPD11066.1 LKAM01000003 KUM49280.1 ASHM01006021 PNY13361.1 OIVN01001114 SPC90048.1 OIVN01002365 SPD03017.1 OIVN01003620 SPD12483.1 OIVN01001336 SPC92838.1 OIVN01003223 SPD09609.1 OIVN01002001 SPC99720.1 OIVN01002225 SPD01801.1 OIVN01006243 SPD28737.1 OIVN01000459 SPC80612.1 OIVN01004260 SPD16354.1 OIVN01006118 SPD25470.1 OIVN01003416 SPD11073.1 OIVN01000817 SPC86004.1 OIVN01001779 OIVN01003965 SPC97547.1 SPD14614.1 OIVN01000702 SPC84305.1 OIVN01000354 SPC78868.1 OIVN01000822 SPC86055.1 CM003613 KYP57192.1 OIVN01002280 SPD02253.1 OIVN01001872 SPC98448.1 OIVN01006344 SPD31070.1 OIVN01000403 SPC79640.1 OIVN01001384 SPC93353.1 OIVN01000452 SPC80504.1 OIVN01001628 SPC96029.1 GEVM01025909 JAU80029.1 OIVN01001171 SPC90720.1 DF237331 GAQ87846.1 CM007895 OTG23274.1 BEGY01000018 GAX76480.1 ASHM01093982 PNX64819.1 DF238519 GAQ93484.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A212F5G1
A0A0A9XV68
A0A0A9XY84
A0A146LH62
W4VRP9
W4VRP2
+ More
A0A0N1ICY1 A0A2N9I4J9 A0A0A9WCG6 A0A2N9ICF5 A0A2N9H9J3 A0A117NI32 A0A2K3PDJ4 A0A2N9FS65 A0A2N9GKN5 A0A2N9HL59 A0A2N9FZX9 A0A2N9HDC0 A0A2N9GJW7 A0A2N9GR48 A0A2N9ITI6 A0A2N9F0H9 A0A2N9HXB8 A0A2N9IMU9 A0A2N9HH69 A0A2N9FGT3 A0A2N9G4F5 A0A2N9FB28 A0A2N9EVG8 A0A2N9FG19 A0A151SR30 A0A2N9GS00 A0A2N9GG65 A0A2N9J3H7 A0A2N9EY13 A0A2N9G1K0 A0A2N9F135 A0A2N9G9I0 A0A1J3IJ96 A0A2N9FUC8 A0A1Y1IGG8 A0A251UJK4 A0A250X157 A0A2K3KEZ8 A0A1Y1IS38
A0A0N1ICY1 A0A2N9I4J9 A0A0A9WCG6 A0A2N9ICF5 A0A2N9H9J3 A0A117NI32 A0A2K3PDJ4 A0A2N9FS65 A0A2N9GKN5 A0A2N9HL59 A0A2N9FZX9 A0A2N9HDC0 A0A2N9GJW7 A0A2N9GR48 A0A2N9ITI6 A0A2N9F0H9 A0A2N9HXB8 A0A2N9IMU9 A0A2N9HH69 A0A2N9FGT3 A0A2N9G4F5 A0A2N9FB28 A0A2N9EVG8 A0A2N9FG19 A0A151SR30 A0A2N9GS00 A0A2N9GG65 A0A2N9J3H7 A0A2N9EY13 A0A2N9G1K0 A0A2N9F135 A0A2N9G9I0 A0A1J3IJ96 A0A2N9FUC8 A0A1Y1IGG8 A0A251UJK4 A0A250X157 A0A2K3KEZ8 A0A1Y1IS38
Ontologies
KEGG
PANTHER
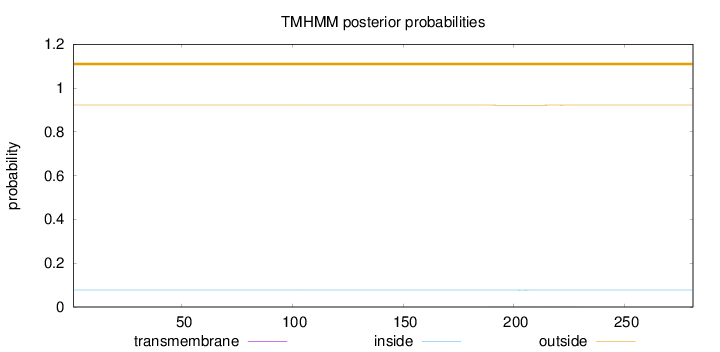
Topology
Length:
281
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0212
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07790
outside
1 - 281
Population Genetic Test Statistics
Pi
5.678937
Theta
7.329747
Tajima's D
-0.818587
CLR
0.801395
CSRT
0.174641267936603
Interpretation
Uncertain
