Gene
KWMTBOMO00952
Pre Gene Modal
BGIBMGA009637
Annotation
PREDICTED:_differentially_expressed_in_FDCP_8_homolog_[Papilio_polytes]
Full name
Differentially expressed in FDCP 8 homolog
Location in the cell
Extracellular Reliability : 1.63 Nuclear Reliability : 1.842
Sequence
CDS
ATGGCAGCGGCTCTCGCTCATGATTCGCCTAAATACCGTGGCAGTTTGATATGCTGTAATCCCAGGACTTCCGTTACGGATCGGAGCCCATCTTCCTCGACTTCTGGTTGCGTGTCTGCTGACGAGAGTTACGACTCAAACTACATACCGAAATCGATAGCGAACAAGAAACTTAAGATACACAGCTCTGCGACCAAAGAAGATATCGAAAAAGCTATAAATCAATGTAAAGAACTAATTTTGAACAGCTCCCAATGCTCCGACGAACGCAAATGGCTAGTGCGTTATTTAGTCGAGCTCAGGCTGAGGTTAGAGGACCTGAAGGACAATGAGGGTCAGACCAGGGTCAAAGTATCCATCAAAGGTCATCACTTCGAGCAGCAGGTCAACCCAAACAACCGCAAACAGTATTGTGATCACTGCAGCGGTGTAATATGGAGTATAGTTCAGAGCTCATATATTTGTACAGACTGTGGATACTTGTGCCATTATAAATGCATCGATCACATATGCAGAGTGTGTGCTCATGTCGTGATGACAGAAAAGGGGGAGTTTGACATGAACATCTGCCCAGAAAGAGGCTTAGCAGCACAAGATTATAAATGTGCTGAATGTGAAACCACATTAACTTTTAAGGATGCTTGGAACGATCCTCGATTATGTGACTATACGGGTCTTTATTACTGTACAACATGCCATTGGAACGATCAGTTTGCGATACCGGCTCGAGTTGTCCACAACTGGGATTGGGAGAAACGTTACGTCTCACGCCTGGCCTACCAAATGTTAAGTATAGCATGGTCACGACCATACATAAACATTGAAAACATTAATTCTAAGCTCTTTAGCTTCATTGCCGAATTGGAATGGGTTCACAAGATGCGCAAGGACCTGGAATGGATGCGTAGATACCTGTGCGCTTGCACCGAAGGCACAACTCTCCTGTCTCCTCTATTCGTCCAACTCGGAGACGTAAACAAGAAGTACAGTATGTCACACTTACAGGCGATTAATGACGGGTCATTAGAAACTGAATTGACTGAACTGACAGAGGTCTGTCGGACACATATAACAAACTGCCCTCTATGTTCAGGCAAAGGTTATTTGTGTGAAGTGTGCAGCAACAATGAAGTGTTGTATCCCTTTGATAGTGGAGCGATAATGTGTGAGAAGTGCAATTCTATGTATCACAGAGGCTGCTGGCTCCGTAAAGGACAGACCTGCCTCAAGTGCACCAGGATAGACGAGAGAAATAAAAGCACCGAAAAGGAAAATGAGAATTTACATTCTGATTATGACATTGACAAATTTGTTGATACCTCTATCGGGCCTCTGAGTGAAACTATCTAG
Protein
MAAALAHDSPKYRGSLICCNPRTSVTDRSPSSSTSGCVSADESYDSNYIPKSIANKKLKIHSSATKEDIEKAINQCKELILNSSQCSDERKWLVRYLVELRLRLEDLKDNEGQTRVKVSIKGHHFEQQVNPNNRKQYCDHCSGVIWSIVQSSYICTDCGYLCHYKCIDHICRVCAHVVMTEKGEFDMNICPERGLAAQDYKCAECETTLTFKDAWNDPRLCDYTGLYYCTTCHWNDQFAIPARVVHNWDWEKRYVSRLAYQMLSIAWSRPYINIENINSKLFSFIAELEWVHKMRKDLEWMRRYLCACTEGTTLLSPLFVQLGDVNKKYSMSHLQAINDGSLETELTELTEVCRTHITNCPLCSGKGYLCEVCSNNEVLYPFDSGAIMCEKCNSMYHRGCWLRKGQTCLKCTRIDERNKSTEKENENLHSDYDIDKFVDTSIGPLSETI
Summary
Similarity
Belongs to the DEF8 family.
Keywords
Complete proteome
Metal-binding
Reference proteome
Repeat
Zinc
Zinc-finger
Feature
chain Differentially expressed in FDCP 8 homolog
Uniprot
H9JJD6
A0A0N1I8V2
A0A194PZR3
A0A2H1W7S8
A0A0L7LLL0
A0A212ET47
+ More
A0A2W1BIV5 A0A2A4J534 A0A1B0FAB8 B4KZ50 A0A1B0AYZ4 A0A1A9UXR3 B4IWM6 A0A0K8TKH1 A0A1I8NBG6 B4LEJ5 A0A0K8V9V2 A0A232FCA6 A0A0A1WWR6 A0A1I8PWW4 W8BEM9 B4GR58 K7IQS8 B5DQN3 A0A0M4F4G5 A0A034W9C1 D6WGG5 A0A3B0J8Q5 A0A0L0BL39 A0A151JPQ1 A0A1B0CQE3 E9J9T1 B4MMN6 A0A1A9W4P8 A0A195FVD4 A0A154PS03 A0A195B5R1 E2BTD4 A0A1I8NBF6 A0A151IJ31 A0A0K8UVZ6 A0A151WGD8 E2ARI0 A0A1I8PX11 A0A0G3Z884 B4QR58 Q9VTT9 B3M7W5 B3NH65 A0A026WQF0 B4PG94 B4HFI8 F4WA97 A0A2J7REP1 A0A1W4UGY3 N6TCR6 U4USD1 A0A1Y1LHQ8 A0A2A3EBB9 A0A088AHU1 A0A0L7RIU1 A0A3L8DQA2 A0A2J7REL2 A0A336LSZ9 A0A067RIK1 B0WIJ1 A0A182IWH1 A0A084WA85 A0A0P4VUF0 W5JEW3 A0A0M9A7A0 A0A0N7ZAP4 R7TQ67 A0A1I8NBH2 A0A182WKR9 A0A182LYG7 A0A182I1Z0 A0A182FCA3 A0A1B6E1E0 A0A182SEE7 K1S180 T1JKT5 A0A182YD36 A0A182RZR4 A0A182PZU5 A0A182X6Z4 A0A182V0K5 V4AU40 A0A182TWQ4 H9GFC1 A0A182JNM4 Q7Q6B8 E0VZS4 A0A2R5L5X8 A0A1B6GU19 A0A293LEX8 F7CTD5 A0A2H6MYR7
A0A2W1BIV5 A0A2A4J534 A0A1B0FAB8 B4KZ50 A0A1B0AYZ4 A0A1A9UXR3 B4IWM6 A0A0K8TKH1 A0A1I8NBG6 B4LEJ5 A0A0K8V9V2 A0A232FCA6 A0A0A1WWR6 A0A1I8PWW4 W8BEM9 B4GR58 K7IQS8 B5DQN3 A0A0M4F4G5 A0A034W9C1 D6WGG5 A0A3B0J8Q5 A0A0L0BL39 A0A151JPQ1 A0A1B0CQE3 E9J9T1 B4MMN6 A0A1A9W4P8 A0A195FVD4 A0A154PS03 A0A195B5R1 E2BTD4 A0A1I8NBF6 A0A151IJ31 A0A0K8UVZ6 A0A151WGD8 E2ARI0 A0A1I8PX11 A0A0G3Z884 B4QR58 Q9VTT9 B3M7W5 B3NH65 A0A026WQF0 B4PG94 B4HFI8 F4WA97 A0A2J7REP1 A0A1W4UGY3 N6TCR6 U4USD1 A0A1Y1LHQ8 A0A2A3EBB9 A0A088AHU1 A0A0L7RIU1 A0A3L8DQA2 A0A2J7REL2 A0A336LSZ9 A0A067RIK1 B0WIJ1 A0A182IWH1 A0A084WA85 A0A0P4VUF0 W5JEW3 A0A0M9A7A0 A0A0N7ZAP4 R7TQ67 A0A1I8NBH2 A0A182WKR9 A0A182LYG7 A0A182I1Z0 A0A182FCA3 A0A1B6E1E0 A0A182SEE7 K1S180 T1JKT5 A0A182YD36 A0A182RZR4 A0A182PZU5 A0A182X6Z4 A0A182V0K5 V4AU40 A0A182TWQ4 H9GFC1 A0A182JNM4 Q7Q6B8 E0VZS4 A0A2R5L5X8 A0A1B6GU19 A0A293LEX8 F7CTD5 A0A2H6MYR7
Pubmed
19121390
26354079
26227816
22118469
28756777
17994087
+ More
26369729 25315136 28648823 25830018 24495485 20075255 15632085 25348373 18362917 19820115 26108605 21282665 20798317 22936249 10731132 12537572 12537569 24508170 17550304 21719571 23537049 28004739 30249741 24845553 24438588 20920257 23761445 23254933 22992520 25244985 21881562 12364791 14747013 17210077 20566863 17495919
26369729 25315136 28648823 25830018 24495485 20075255 15632085 25348373 18362917 19820115 26108605 21282665 20798317 22936249 10731132 12537572 12537569 24508170 17550304 21719571 23537049 28004739 30249741 24845553 24438588 20920257 23761445 23254933 22992520 25244985 21881562 12364791 14747013 17210077 20566863 17495919
EMBL
BABH01017870
KQ460296
KPJ16274.1
KQ459593
KPI96655.1
ODYU01006575
+ More
SOQ48534.1 JTDY01000650 KOB76332.1 AGBW02012654 OWR44649.1 KZ150094 PZC73635.1 NWSH01003281 PCG66624.1 CCAG010015091 CH933809 EDW17847.1 JXJN01006061 CH916366 EDV96252.1 GDAI01002751 JAI14852.1 CH940647 EDW69080.1 GDHF01016672 JAI35642.1 NNAY01000475 OXU28120.1 GBXI01011459 JAD02833.1 GAMC01006795 JAB99760.1 CH479188 EDW40243.1 CH379069 EDY73456.1 CP012526 ALC46270.1 GAKP01007653 JAC51299.1 KQ971321 EFA00553.2 OUUW01000002 SPP76262.1 JRES01001708 KNC20663.1 KQ978756 KYN28584.1 AJWK01023464 GL769399 EFZ10461.1 CH963847 EDW73442.1 KQ981272 KYN43824.1 KQ435119 KZC14705.1 KQ976595 KYM79610.1 GL450389 EFN81055.1 KQ977412 KYN02862.1 GDHF01021573 JAI30741.1 KQ983176 KYQ46913.1 GL442063 EFN63989.1 KP247436 AKM45842.1 CM000363 CM002912 EDX10188.1 KMY99165.1 AE014296 AY058352 CH902618 EDV38838.1 CH954178 EDV51522.1 KK107128 EZA58245.1 CM000159 EDW94258.1 CH480815 EDW41219.1 GL888045 EGI68888.1 NEVH01004959 PNF39277.1 APGK01042943 KB741011 ENN75518.1 KB632341 ERL93005.1 GEZM01057190 JAV72398.1 KZ288310 PBC28592.1 KQ414583 KOC70728.1 QOIP01000006 RLU22089.1 PNF39278.1 UFQS01000156 UFQT01000156 SSX00625.1 SSX21005.1 KK852482 KDR22863.1 DS231949 EDS28514.1 ATLV01022053 KE525327 KFB47129.1 GDRN01096078 JAI59412.1 ADMH02001414 ETN62626.1 KQ435727 KOX77916.1 GDRN01096076 GDRN01096073 JAI59416.1 AMQN01011671 KB309013 ELT95789.1 AXCM01008814 APCN01000100 GEDC01021489 GEDC01005556 JAS15809.1 JAS31742.1 JH823218 EKC41006.1 JH431796 AXCN02000418 KB201305 ESO97291.1 AAWZ02033487 AAAB01008960 EAA11791.4 DS235854 EEB18880.1 GGLE01000772 MBY04898.1 GECZ01003853 JAS65916.1 GFWV01002563 MAA27293.1 IACI01011223 LAA20463.1
SOQ48534.1 JTDY01000650 KOB76332.1 AGBW02012654 OWR44649.1 KZ150094 PZC73635.1 NWSH01003281 PCG66624.1 CCAG010015091 CH933809 EDW17847.1 JXJN01006061 CH916366 EDV96252.1 GDAI01002751 JAI14852.1 CH940647 EDW69080.1 GDHF01016672 JAI35642.1 NNAY01000475 OXU28120.1 GBXI01011459 JAD02833.1 GAMC01006795 JAB99760.1 CH479188 EDW40243.1 CH379069 EDY73456.1 CP012526 ALC46270.1 GAKP01007653 JAC51299.1 KQ971321 EFA00553.2 OUUW01000002 SPP76262.1 JRES01001708 KNC20663.1 KQ978756 KYN28584.1 AJWK01023464 GL769399 EFZ10461.1 CH963847 EDW73442.1 KQ981272 KYN43824.1 KQ435119 KZC14705.1 KQ976595 KYM79610.1 GL450389 EFN81055.1 KQ977412 KYN02862.1 GDHF01021573 JAI30741.1 KQ983176 KYQ46913.1 GL442063 EFN63989.1 KP247436 AKM45842.1 CM000363 CM002912 EDX10188.1 KMY99165.1 AE014296 AY058352 CH902618 EDV38838.1 CH954178 EDV51522.1 KK107128 EZA58245.1 CM000159 EDW94258.1 CH480815 EDW41219.1 GL888045 EGI68888.1 NEVH01004959 PNF39277.1 APGK01042943 KB741011 ENN75518.1 KB632341 ERL93005.1 GEZM01057190 JAV72398.1 KZ288310 PBC28592.1 KQ414583 KOC70728.1 QOIP01000006 RLU22089.1 PNF39278.1 UFQS01000156 UFQT01000156 SSX00625.1 SSX21005.1 KK852482 KDR22863.1 DS231949 EDS28514.1 ATLV01022053 KE525327 KFB47129.1 GDRN01096078 JAI59412.1 ADMH02001414 ETN62626.1 KQ435727 KOX77916.1 GDRN01096076 GDRN01096073 JAI59416.1 AMQN01011671 KB309013 ELT95789.1 AXCM01008814 APCN01000100 GEDC01021489 GEDC01005556 JAS15809.1 JAS31742.1 JH823218 EKC41006.1 JH431796 AXCN02000418 KB201305 ESO97291.1 AAWZ02033487 AAAB01008960 EAA11791.4 DS235854 EEB18880.1 GGLE01000772 MBY04898.1 GECZ01003853 JAS65916.1 GFWV01002563 MAA27293.1 IACI01011223 LAA20463.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000007151
UP000218220
+ More
UP000092444 UP000009192 UP000092460 UP000078200 UP000001070 UP000095301 UP000008792 UP000215335 UP000095300 UP000008744 UP000002358 UP000001819 UP000092553 UP000007266 UP000268350 UP000037069 UP000078492 UP000092461 UP000007798 UP000091820 UP000078541 UP000076502 UP000078540 UP000008237 UP000078542 UP000075809 UP000000311 UP000000304 UP000000803 UP000007801 UP000008711 UP000053097 UP000002282 UP000001292 UP000007755 UP000235965 UP000192221 UP000019118 UP000030742 UP000242457 UP000005203 UP000053825 UP000279307 UP000027135 UP000002320 UP000075880 UP000030765 UP000000673 UP000053105 UP000014760 UP000075920 UP000075883 UP000075840 UP000069272 UP000075901 UP000005408 UP000076408 UP000075900 UP000075886 UP000076407 UP000075903 UP000030746 UP000075902 UP000001646 UP000075881 UP000007062 UP000009046 UP000002280
UP000092444 UP000009192 UP000092460 UP000078200 UP000001070 UP000095301 UP000008792 UP000215335 UP000095300 UP000008744 UP000002358 UP000001819 UP000092553 UP000007266 UP000268350 UP000037069 UP000078492 UP000092461 UP000007798 UP000091820 UP000078541 UP000076502 UP000078540 UP000008237 UP000078542 UP000075809 UP000000311 UP000000304 UP000000803 UP000007801 UP000008711 UP000053097 UP000002282 UP000001292 UP000007755 UP000235965 UP000192221 UP000019118 UP000030742 UP000242457 UP000005203 UP000053825 UP000279307 UP000027135 UP000002320 UP000075880 UP000030765 UP000000673 UP000053105 UP000014760 UP000075920 UP000075883 UP000075840 UP000069272 UP000075901 UP000005408 UP000076408 UP000075900 UP000075886 UP000076407 UP000075903 UP000030746 UP000075902 UP000001646 UP000075881 UP000007062 UP000009046 UP000002280
Interpro
SUPFAM
SSF48695
SSF48695
CDD
ProteinModelPortal
H9JJD6
A0A0N1I8V2
A0A194PZR3
A0A2H1W7S8
A0A0L7LLL0
A0A212ET47
+ More
A0A2W1BIV5 A0A2A4J534 A0A1B0FAB8 B4KZ50 A0A1B0AYZ4 A0A1A9UXR3 B4IWM6 A0A0K8TKH1 A0A1I8NBG6 B4LEJ5 A0A0K8V9V2 A0A232FCA6 A0A0A1WWR6 A0A1I8PWW4 W8BEM9 B4GR58 K7IQS8 B5DQN3 A0A0M4F4G5 A0A034W9C1 D6WGG5 A0A3B0J8Q5 A0A0L0BL39 A0A151JPQ1 A0A1B0CQE3 E9J9T1 B4MMN6 A0A1A9W4P8 A0A195FVD4 A0A154PS03 A0A195B5R1 E2BTD4 A0A1I8NBF6 A0A151IJ31 A0A0K8UVZ6 A0A151WGD8 E2ARI0 A0A1I8PX11 A0A0G3Z884 B4QR58 Q9VTT9 B3M7W5 B3NH65 A0A026WQF0 B4PG94 B4HFI8 F4WA97 A0A2J7REP1 A0A1W4UGY3 N6TCR6 U4USD1 A0A1Y1LHQ8 A0A2A3EBB9 A0A088AHU1 A0A0L7RIU1 A0A3L8DQA2 A0A2J7REL2 A0A336LSZ9 A0A067RIK1 B0WIJ1 A0A182IWH1 A0A084WA85 A0A0P4VUF0 W5JEW3 A0A0M9A7A0 A0A0N7ZAP4 R7TQ67 A0A1I8NBH2 A0A182WKR9 A0A182LYG7 A0A182I1Z0 A0A182FCA3 A0A1B6E1E0 A0A182SEE7 K1S180 T1JKT5 A0A182YD36 A0A182RZR4 A0A182PZU5 A0A182X6Z4 A0A182V0K5 V4AU40 A0A182TWQ4 H9GFC1 A0A182JNM4 Q7Q6B8 E0VZS4 A0A2R5L5X8 A0A1B6GU19 A0A293LEX8 F7CTD5 A0A2H6MYR7
A0A2W1BIV5 A0A2A4J534 A0A1B0FAB8 B4KZ50 A0A1B0AYZ4 A0A1A9UXR3 B4IWM6 A0A0K8TKH1 A0A1I8NBG6 B4LEJ5 A0A0K8V9V2 A0A232FCA6 A0A0A1WWR6 A0A1I8PWW4 W8BEM9 B4GR58 K7IQS8 B5DQN3 A0A0M4F4G5 A0A034W9C1 D6WGG5 A0A3B0J8Q5 A0A0L0BL39 A0A151JPQ1 A0A1B0CQE3 E9J9T1 B4MMN6 A0A1A9W4P8 A0A195FVD4 A0A154PS03 A0A195B5R1 E2BTD4 A0A1I8NBF6 A0A151IJ31 A0A0K8UVZ6 A0A151WGD8 E2ARI0 A0A1I8PX11 A0A0G3Z884 B4QR58 Q9VTT9 B3M7W5 B3NH65 A0A026WQF0 B4PG94 B4HFI8 F4WA97 A0A2J7REP1 A0A1W4UGY3 N6TCR6 U4USD1 A0A1Y1LHQ8 A0A2A3EBB9 A0A088AHU1 A0A0L7RIU1 A0A3L8DQA2 A0A2J7REL2 A0A336LSZ9 A0A067RIK1 B0WIJ1 A0A182IWH1 A0A084WA85 A0A0P4VUF0 W5JEW3 A0A0M9A7A0 A0A0N7ZAP4 R7TQ67 A0A1I8NBH2 A0A182WKR9 A0A182LYG7 A0A182I1Z0 A0A182FCA3 A0A1B6E1E0 A0A182SEE7 K1S180 T1JKT5 A0A182YD36 A0A182RZR4 A0A182PZU5 A0A182X6Z4 A0A182V0K5 V4AU40 A0A182TWQ4 H9GFC1 A0A182JNM4 Q7Q6B8 E0VZS4 A0A2R5L5X8 A0A1B6GU19 A0A293LEX8 F7CTD5 A0A2H6MYR7
PDB
4L9M
E-value=4.14503e-05,
Score=113
Ontologies
PANTHER
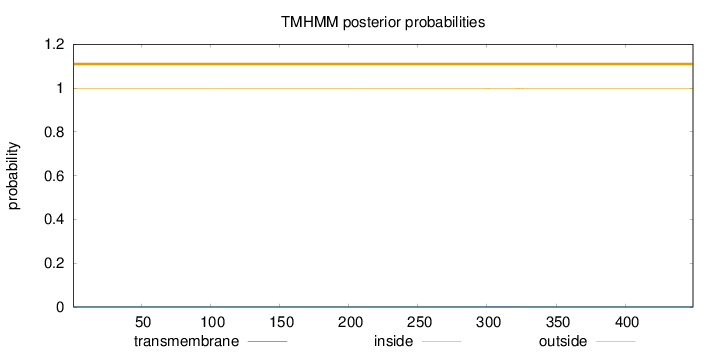
Topology
Length:
449
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02436
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00259
outside
1 - 449
Population Genetic Test Statistics
Pi
323.95917
Theta
224.842633
Tajima's D
1.388177
CLR
0
CSRT
0.75851207439628
Interpretation
Uncertain
