Gene
KWMTBOMO00941
Pre Gene Modal
BGIBMGA009641
Annotation
peritrophin_type-A_domain_protein_1_[Mamestra_configurata]
Location in the cell
Extracellular Reliability : 3.356
Sequence
CDS
ATGCTATGCAATAGATTTACCGCGGCAGTCCCGGAGTGCCGCACGTGTTTTTTCCGTACCTATACGCTGATAGGTAAAGCCCTTAGTCTCTTGCTGTTGACCGGCGTCGTGTCCTGTGCTGAAAACGCTACAGAGAGCGTGTTCAGGACTTTTAACAAAGTCGATCCCAACTCACTGTCGTGTGACCCAGCGGGACATGTCTTCCTGCTTCTACCACACTTCCGTGACTGCTCGAAGTTCTACATGTGCGCTCACGGGGAAGAAGTGGAATTCAATTGCAACGGCGGCTTGATATTCGATTTCGAGTTGCAGACATGCAACTGGAAATGGGCTACGAATTGTACCCTTCGTGTTGTGAAAGAGGACATCGACACTGAAGGTTCGGGCTTGGGTTCCGGGGAAGAGACCATCGGTATATTTGGTGAAGAAGTAGAAAACGGACCTATAGATATCCTCACAGCAGACTCAGCAGGCACAGTGAGGCCGCTGTCTCCAAGTAATTTGAGGTTCTTCAACCACGAGCTGAACTGCCACAGGGCTGATGCTGCAGCGAAACAAGTGGCTTACAAGGGCGATTGTCAGCGGTACTGGCGGTGCGTGGGCGGCGTGCCTCAAGCCATGTACTGTACGGATGGACTCTTCTTCAACGAGCTGACGCAGCAGTGTGACTTCGAGGCCAACGTCAAGTGCGGCGTGATACCGGACGAGGAGCTACAAGGCGAATTCATTGTGTACAAAAACTAG
Protein
MLCNRFTAAVPECRTCFFRTYTLIGKALSLLLLTGVVSCAENATESVFRTFNKVDPNSLSCDPAGHVFLLLPHFRDCSKFYMCAHGEEVEFNCNGGLIFDFELQTCNWKWATNCTLRVVKEDIDTEGSGLGSGEETIGIFGEEVENGPIDILTADSAGTVRPLSPSNLRFFNHELNCHRADAAAKQVAYKGDCQRYWRCVGGVPQAMYCTDGLFFNELTQQCDFEANVKCGVIPDEELQGEFIVYKN
Summary
Uniprot
A0A0F6QDS3
H9JJE0
D5KU16
B6CMG4
A0A2H1WH58
A0A194PZQ3
+ More
A0A0N1I8W5 B8ZX09 A0A212F4S2 A0A212ERD9 A0A1E1W2G1 D1MAJ8 A0A2H1V8J7 A0A2A4JPM5 Q86BV0 A0A0L7LSJ1 Q6VAP0 B6CAP5 Q6VAN9 A0A194PZ93 A0A2A4JFF0 A0A2A4JDW5 A0A2A4JEA8 A0A2H1WHD8 I3RQC1 L7X1F1 A0A194RTC3 C4N8D2 C4N881 A0A194RPW9 A0A0Q9WJF7 A0A2A4JAZ3 A0A088BU36 C4N882 E7D2J6 T1IW66 W5JA06 M1EYJ2 A0A2M4DRY4 B5DRI4 B4H8N3 B4H8N2 Q29DL6 A0A0L7KUX8 A0A3M7R512 A0A194PZ97 A0A3B0JLT1 B3NHF6 A0A3B0JRQ1 B4IXM2 B4ITJ6 A0A0C9PYS6 A0A087TZ26 A0A0C9RID9 A0A0M3QWL7 B3NHF5 A0A0J9RUM5 B4QJ31
A0A0N1I8W5 B8ZX09 A0A212F4S2 A0A212ERD9 A0A1E1W2G1 D1MAJ8 A0A2H1V8J7 A0A2A4JPM5 Q86BV0 A0A0L7LSJ1 Q6VAP0 B6CAP5 Q6VAN9 A0A194PZ93 A0A2A4JFF0 A0A2A4JDW5 A0A2A4JEA8 A0A2H1WHD8 I3RQC1 L7X1F1 A0A194RTC3 C4N8D2 C4N881 A0A194RPW9 A0A0Q9WJF7 A0A2A4JAZ3 A0A088BU36 C4N882 E7D2J6 T1IW66 W5JA06 M1EYJ2 A0A2M4DRY4 B5DRI4 B4H8N3 B4H8N2 Q29DL6 A0A0L7KUX8 A0A3M7R512 A0A194PZ97 A0A3B0JLT1 B3NHF6 A0A3B0JRQ1 B4IXM2 B4ITJ6 A0A0C9PYS6 A0A087TZ26 A0A0C9RID9 A0A0M3QWL7 B3NHF5 A0A0J9RUM5 B4QJ31
Pubmed
EMBL
KP744646
AKD49097.1
BABH01017856
GU596430
ADE06396.1
EU325564
+ More
ACB54954.1 ODYU01008643 SOQ52398.1 KQ459593 KPI96645.1 KQ460280 KPJ16303.1 FM955425 CAW30926.1 AGBW02010314 OWR48745.1 AGBW02013028 OWR44066.1 GDQN01009887 JAT81167.1 GU128106 KQ971357 ACY95489.1 KYB26070.1 ODYU01001235 SOQ37170.1 NWSH01000891 PCG73658.1 AY277403 AAP33177.1 JTDY01000178 KOB78427.1 AY345124 AAR06265.1 EU139126 ABW98673.1 AY345125 AAR06266.1 KQ459585 KPI98363.1 NWSH01001780 PCG70160.1 PCG70161.1 PCG70159.1 ODYU01008691 SOQ52478.1 JQ730735 AFK27934.1 KC108816 AGC94490.1 KQ459896 KPJ19356.1 FJ408730 ACQ65651.1 FJ393548 ACR20468.1 KPJ19360.1 CH940647 KRF84691.1 NWSH01002232 PCG68868.1 KC703970 AHH02588.1 FJ393549 ACR20469.1 HQ012005 ADV03161.1 JH431608 ADMH02002072 ETN59594.1 JF681185 AFD28281.1 GGFL01016138 MBW80316.1 CH379070 EDY74171.1 CH479224 EDW35076.1 EDW35075.1 EAL30398.3 JTDY01005617 KOB66824.1 REGN01004195 RNA18636.1 KPI98368.1 OUUW01000002 SPP76380.1 CH954178 EDV51682.1 SPP76379.1 CH916366 EDV97484.1 CH891711 EDW99709.1 GBYB01006678 JAG76445.1 KK117393 KFM70365.1 GBYB01006676 JAG76443.1 CP012525 ALC44369.1 EDV51681.1 CM002912 KMY99398.1 CM000363 EDX10362.1 KMY99397.1
ACB54954.1 ODYU01008643 SOQ52398.1 KQ459593 KPI96645.1 KQ460280 KPJ16303.1 FM955425 CAW30926.1 AGBW02010314 OWR48745.1 AGBW02013028 OWR44066.1 GDQN01009887 JAT81167.1 GU128106 KQ971357 ACY95489.1 KYB26070.1 ODYU01001235 SOQ37170.1 NWSH01000891 PCG73658.1 AY277403 AAP33177.1 JTDY01000178 KOB78427.1 AY345124 AAR06265.1 EU139126 ABW98673.1 AY345125 AAR06266.1 KQ459585 KPI98363.1 NWSH01001780 PCG70160.1 PCG70161.1 PCG70159.1 ODYU01008691 SOQ52478.1 JQ730735 AFK27934.1 KC108816 AGC94490.1 KQ459896 KPJ19356.1 FJ408730 ACQ65651.1 FJ393548 ACR20468.1 KPJ19360.1 CH940647 KRF84691.1 NWSH01002232 PCG68868.1 KC703970 AHH02588.1 FJ393549 ACR20469.1 HQ012005 ADV03161.1 JH431608 ADMH02002072 ETN59594.1 JF681185 AFD28281.1 GGFL01016138 MBW80316.1 CH379070 EDY74171.1 CH479224 EDW35076.1 EDW35075.1 EAL30398.3 JTDY01005617 KOB66824.1 REGN01004195 RNA18636.1 KPI98368.1 OUUW01000002 SPP76380.1 CH954178 EDV51682.1 SPP76379.1 CH916366 EDV97484.1 CH891711 EDW99709.1 GBYB01006678 JAG76445.1 KK117393 KFM70365.1 GBYB01006676 JAG76443.1 CP012525 ALC44369.1 EDV51681.1 CM002912 KMY99398.1 CM000363 EDX10362.1 KMY99397.1
Proteomes
PRIDE
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
A0A0F6QDS3
H9JJE0
D5KU16
B6CMG4
A0A2H1WH58
A0A194PZQ3
+ More
A0A0N1I8W5 B8ZX09 A0A212F4S2 A0A212ERD9 A0A1E1W2G1 D1MAJ8 A0A2H1V8J7 A0A2A4JPM5 Q86BV0 A0A0L7LSJ1 Q6VAP0 B6CAP5 Q6VAN9 A0A194PZ93 A0A2A4JFF0 A0A2A4JDW5 A0A2A4JEA8 A0A2H1WHD8 I3RQC1 L7X1F1 A0A194RTC3 C4N8D2 C4N881 A0A194RPW9 A0A0Q9WJF7 A0A2A4JAZ3 A0A088BU36 C4N882 E7D2J6 T1IW66 W5JA06 M1EYJ2 A0A2M4DRY4 B5DRI4 B4H8N3 B4H8N2 Q29DL6 A0A0L7KUX8 A0A3M7R512 A0A194PZ97 A0A3B0JLT1 B3NHF6 A0A3B0JRQ1 B4IXM2 B4ITJ6 A0A0C9PYS6 A0A087TZ26 A0A0C9RID9 A0A0M3QWL7 B3NHF5 A0A0J9RUM5 B4QJ31
A0A0N1I8W5 B8ZX09 A0A212F4S2 A0A212ERD9 A0A1E1W2G1 D1MAJ8 A0A2H1V8J7 A0A2A4JPM5 Q86BV0 A0A0L7LSJ1 Q6VAP0 B6CAP5 Q6VAN9 A0A194PZ93 A0A2A4JFF0 A0A2A4JDW5 A0A2A4JEA8 A0A2H1WHD8 I3RQC1 L7X1F1 A0A194RTC3 C4N8D2 C4N881 A0A194RPW9 A0A0Q9WJF7 A0A2A4JAZ3 A0A088BU36 C4N882 E7D2J6 T1IW66 W5JA06 M1EYJ2 A0A2M4DRY4 B5DRI4 B4H8N3 B4H8N2 Q29DL6 A0A0L7KUX8 A0A3M7R512 A0A194PZ97 A0A3B0JLT1 B3NHF6 A0A3B0JRQ1 B4IXM2 B4ITJ6 A0A0C9PYS6 A0A087TZ26 A0A0C9RID9 A0A0M3QWL7 B3NHF5 A0A0J9RUM5 B4QJ31
PDB
6G9E
E-value=0.000248467,
Score=103
Ontologies
GO
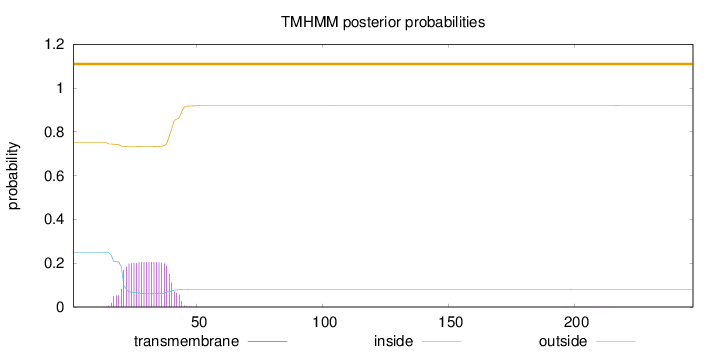
Topology
Length:
247
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.35434
Exp number, first 60 AAs:
4.34838
Total prob of N-in:
0.24938
outside
1 - 247
Population Genetic Test Statistics
Pi
212.297394
Theta
138.826418
Tajima's D
1.422798
CLR
0.009154
CSRT
0.769511524423779
Interpretation
Uncertain
