Gene
KWMTBOMO00938 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009799
Annotation
PREDICTED:_aldo-keto_reductase_AKR2E4-like_isoform_X2_[Bombyx_mori]
Full name
Aldo-keto reductase AKR2E4
Alternative Name
3-dehydroecdysone reductase
Aldo-keto reductase 2E
Aldo-keto reductase 2E
Location in the cell
Cytoplasmic Reliability : 3.55
Sequence
CDS
ATGGCTAAGGTATCGATTCCAACGATTAAATTGAACAATGGAATCGAATTCCCGGCCCTCGGCTACGGGACCTGGCTAGGACTGAACGATAAGATGGAATTCGAAATCACGGACGCGCGTAAACTACAGGACGCATTATCGTACGCCATCGATATAGGTTATCGACATATCGACACGGCGCATCTCTACCGAGTCGAGCCGGAGGTCGGGGAAGTTATAAATAAAAAAATTAAGGACGGTGTTGTCAAACGCGAAGACATTTTTGTTACTACAAAGGTGTGGCACCACCACCACAAGCAAGCGGACGTCGAGGTGGCGGTGCGCGACTGCCTCAGGAGACTGAATCTGGAGTACGTGGACTTGGTATTGGTGCACTGGCCAATGTCGATCTCGCAAGAAGGGGTAGACGAAAATATCGACTACCTGGAGACGTGGAGAGGGTTTGAGGAGGTCCTCAAGAAGGGACTGACCAGGTCCATCGGCGTGTCCAACTTCAACATCGAACAGATGAAGAGGTTACTAGCCAACTGCAACGTGCCGCCCGCCGTCAACCAGATCGAGATCAACCTAAATCTCGGTCAGAAGGAATTGGTGGACTACTGTCAGTCCAAGGGCGTGGTGGTGGTCGCGTACTCGCCATTCGGAACGATGGTGCCGTCCAGAAACTACCCGGACAGTCCGGAGCCTAAGATGGACAATCCCACGATGCTCTCGATAGCGAAGAAACACGGGAAGACCGTCACTCAGATCGCCTTAAGATATTTGTACCAGCGCGGGATCGTGTCTATACCGAAGACTGTGACTAAGAGCAGAGTGCTGGAGAACGCCTCGATCTTCGACTTCCAGCTGGACGACGAGGACGTCGCCACCCTGGCGCAGTTCGACAACGGCTTCCGGACGATCAGGCCGCTGTTCTGGCAGCCCTACGAGAACTACCCCTTCGATGTGGTCCTTGGACAAGAGCACGAAGACATACCGATCTCTTTGCTCAAGTGGAAGAATGGAGTCAACGGCGACATCGAATAA
Protein
MAKVSIPTIKLNNGIEFPALGYGTWLGLNDKMEFEITDARKLQDALSYAIDIGYRHIDTAHLYRVEPEVGEVINKKIKDGVVKREDIFVTTKVWHHHHKQADVEVAVRDCLRRLNLEYVDLVLVHWPMSISQEGVDENIDYLETWRGFEEVLKKGLTRSIGVSNFNIEQMKRLLANCNVPPAVNQIEINLNLGQKELVDYCQSKGVVVVAYSPFGTMVPSRNYPDSPEPKMDNPTMLSIAKKHGKTVTQIALRYLYQRGIVSIPKTVTKSRVLENASIFDFQLDDEDVATLAQFDNGFRTIRPLFWQPYENYPFDVVLGQEHEDIPISLLKWKNGVNGDIE
Summary
Description
NADP-dependent oxidoreductase with high 3-dehydroecdysone reductase activity. May play a role in the regulation of molting. Has lower activity with phenylglyoxal and isatin (in vitro). Has no activity with NADH as cosubstrate. Has no activity with nitrobenzaldehyde and 3-hydroxybenzaldehyde.
Biophysicochemical Properties
0.0044 mM for 3-dehydroecdysone
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
3D-structure
Complete proteome
Lipid metabolism
NADP
Oxidoreductase
Reference proteome
Steroid metabolism
Feature
chain Aldo-keto reductase AKR2E4
Uniprot
H9JJU8
H9JJU9
A0A0H5AQ19
B8ZX07
A0A291P0V2
A0A291P0V4
+ More
G9F9G1 I4DKC0 A0A0N1PGT3 A0A212ENH0 U5KCB5 A0A194PV62 A0A2A4J392 A0A212ERG2 A0A2A4J1K6 H9IVT6 I4DKQ3 A0A291P0T7 A0A2A4JMB2 A0A1L8D6L9 A0A194RJ39 A0A2A4K2R0 Q9Y020 A0A2A4IW06 A0A291P0Y7 A0A1B2G267 A0A1L8D6N3 A0A1Z2RRK0 A0A2W1BLQ6 U5KCB8 A0A2W1BC51 G9F9G0 A0A194Q090 A0A194PKB3 I4DP16 D2SNM4 A0A2H1WKT0 A0A291P0X3 A0A2H1VAA3 U5KBY4 A0A212FE35 H9IVR7 A0A194R6Z8 A0A291P0W7 A0A212EU63 H9IVS0 A0A291P0U2 V5GQ24 A0A194PSF7 A0A291P0Y9 G9F9F4 A0A291P0S0 A0A0C9QAV8 A0A212FE89 A0A2A4IYA3 D2SNV7 G9F9F8 A0A232EMX7 A0A194PUL9 A0A067QJF4 A0A0F6Q2W6 A0A212FML6 F4WLB4 A0A0C9R605 J3JX35 A0A2H1WKV4 A0A1D2MB39 H9JTG9 G9F9F7 A0A0U4B4X4 A0A0L7QVJ2 A0A212F244 K7IXG3 A0A1W4X5Y5 V5GT06 A0A154PH71 A0A195CTD6 A0A182QZ12 A0A088AGI5 A0A1L8D6N6 E9J719 V4B296 A0A291P0Z6 A0A310SCS5 V4AZ56 N6UG67 A0A2A3EI70 V9IEZ4 A0A1I9WLL7 A0A2H1V815 A0A1L8E8F0 S4P5J6 A0A1L8E7Z6 A0A1L8E836 A0A291P0V1 A0A182M9D3
G9F9G1 I4DKC0 A0A0N1PGT3 A0A212ENH0 U5KCB5 A0A194PV62 A0A2A4J392 A0A212ERG2 A0A2A4J1K6 H9IVT6 I4DKQ3 A0A291P0T7 A0A2A4JMB2 A0A1L8D6L9 A0A194RJ39 A0A2A4K2R0 Q9Y020 A0A2A4IW06 A0A291P0Y7 A0A1B2G267 A0A1L8D6N3 A0A1Z2RRK0 A0A2W1BLQ6 U5KCB8 A0A2W1BC51 G9F9G0 A0A194Q090 A0A194PKB3 I4DP16 D2SNM4 A0A2H1WKT0 A0A291P0X3 A0A2H1VAA3 U5KBY4 A0A212FE35 H9IVR7 A0A194R6Z8 A0A291P0W7 A0A212EU63 H9IVS0 A0A291P0U2 V5GQ24 A0A194PSF7 A0A291P0Y9 G9F9F4 A0A291P0S0 A0A0C9QAV8 A0A212FE89 A0A2A4IYA3 D2SNV7 G9F9F8 A0A232EMX7 A0A194PUL9 A0A067QJF4 A0A0F6Q2W6 A0A212FML6 F4WLB4 A0A0C9R605 J3JX35 A0A2H1WKV4 A0A1D2MB39 H9JTG9 G9F9F7 A0A0U4B4X4 A0A0L7QVJ2 A0A212F244 K7IXG3 A0A1W4X5Y5 V5GT06 A0A154PH71 A0A195CTD6 A0A182QZ12 A0A088AGI5 A0A1L8D6N6 E9J719 V4B296 A0A291P0Z6 A0A310SCS5 V4AZ56 N6UG67 A0A2A3EI70 V9IEZ4 A0A1I9WLL7 A0A2H1V815 A0A1L8E8F0 S4P5J6 A0A1L8E7Z6 A0A1L8E836 A0A291P0V1 A0A182M9D3
EC Number
1.1.1.-
Pubmed
EMBL
BABH01017849
BABH01017850
BABH01017851
BABH01017852
LC012974
LC375688
+ More
BAR91212.1 BBD35023.1 FM955425 CAW30924.1 MF687611 ATJ44537.1 MF687578 ATJ44504.1 JN033706 AEW46859.1 AK401738 BAM18360.1 KQ460280 KPJ16305.1 AGBW02013667 OWR43007.1 KC007346 AGQ45611.1 KQ459593 KPI96644.1 NWSH01003732 PCG65872.1 AGBW02013028 OWR44065.1 PCG65871.1 BABH01038068 BABH01038069 BABH01038070 AK401871 BAM18493.1 MF687576 ATJ44502.1 NWSH01001060 PCG72858.1 GEYN01000020 JAV02109.1 KQ460152 KPJ17355.1 NWSH01000243 PCG78063.1 AJ131966 CAB41997.1 NWSH01006101 PCG63656.1 MF687609 ATJ44535.1 KU233524 ANZ02995.1 GEYN01000016 JAV02113.1 KY938815 ASA46456.1 KZ150094 PZC73640.1 KC007356 AGQ45621.1 KZ150230 PZC72001.1 JN033705 AEW46858.1 KQ459582 KPI98997.1 KQ459602 KPI93169.1 AK403246 BAM19656.1 EZ407153 ACX53715.1 ODYU01009309 SOQ53638.1 MF687617 ATJ44543.1 ODYU01001496 SOQ37780.1 KC007350 AGQ45615.1 AGBW02008974 OWR52019.1 BABH01038087 BABH01038088 BABH01038089 BABH01038090 KQ460878 KPJ11641.1 MF687615 ATJ44541.1 AGBW02012481 OWR44997.1 BABH01038066 LC375687 BBD35022.1 MF687572 ATJ44498.1 GALX01004754 JAB63712.1 KPI96371.1 MF687608 ATJ44534.1 JN033699 JN033700 RSAL01000051 AEW46852.1 AEW46853.1 RVE50216.1 MF687575 ATJ44501.1 GBYB01000353 GBYB01000354 JAG70120.1 JAG70121.1 OWR52018.1 NWSH01005088 PCG64446.1 EZ407241 ACX53798.1 JN033703 AEW46856.1 NNAY01003257 OXU19723.1 KPI96444.1 KK853419 KDR07716.1 KP237872 AKD01725.1 AGBW02007651 OWR54929.1 GL888207 EGI65077.1 GBYB01002216 JAG71983.1 BT127803 AEE62765.1 SOQ53637.1 LJIJ01002106 ODM90218.1 AB701383 JN033702 AEW46855.1 KU218649 ALX00029.1 KQ414726 KOC62648.1 AGBW02010783 OWR47810.1 AAZX01000044 GALX01003699 JAB64767.1 KQ434902 KZC11162.1 KQ977349 KYN03394.1 AXCN02001099 GEYN01000021 JAV02108.1 GL768421 EFZ11357.1 KB200718 ESP00397.1 MF687618 ATJ44544.1 KQ777390 OAD52083.1 ESP00396.1 APGK01021961 APGK01021962 KB740392 ENN80755.1 KZ288235 PBC31398.1 JR040676 AEY59237.1 KU932401 APA34037.1 ODYU01001161 SOQ36985.1 GFDG01003937 JAV14862.1 GAIX01005504 JAA87056.1 GFDG01003939 JAV14860.1 GFDG01003938 JAV14861.1 MF687580 ATJ44506.1 AXCM01001195
BAR91212.1 BBD35023.1 FM955425 CAW30924.1 MF687611 ATJ44537.1 MF687578 ATJ44504.1 JN033706 AEW46859.1 AK401738 BAM18360.1 KQ460280 KPJ16305.1 AGBW02013667 OWR43007.1 KC007346 AGQ45611.1 KQ459593 KPI96644.1 NWSH01003732 PCG65872.1 AGBW02013028 OWR44065.1 PCG65871.1 BABH01038068 BABH01038069 BABH01038070 AK401871 BAM18493.1 MF687576 ATJ44502.1 NWSH01001060 PCG72858.1 GEYN01000020 JAV02109.1 KQ460152 KPJ17355.1 NWSH01000243 PCG78063.1 AJ131966 CAB41997.1 NWSH01006101 PCG63656.1 MF687609 ATJ44535.1 KU233524 ANZ02995.1 GEYN01000016 JAV02113.1 KY938815 ASA46456.1 KZ150094 PZC73640.1 KC007356 AGQ45621.1 KZ150230 PZC72001.1 JN033705 AEW46858.1 KQ459582 KPI98997.1 KQ459602 KPI93169.1 AK403246 BAM19656.1 EZ407153 ACX53715.1 ODYU01009309 SOQ53638.1 MF687617 ATJ44543.1 ODYU01001496 SOQ37780.1 KC007350 AGQ45615.1 AGBW02008974 OWR52019.1 BABH01038087 BABH01038088 BABH01038089 BABH01038090 KQ460878 KPJ11641.1 MF687615 ATJ44541.1 AGBW02012481 OWR44997.1 BABH01038066 LC375687 BBD35022.1 MF687572 ATJ44498.1 GALX01004754 JAB63712.1 KPI96371.1 MF687608 ATJ44534.1 JN033699 JN033700 RSAL01000051 AEW46852.1 AEW46853.1 RVE50216.1 MF687575 ATJ44501.1 GBYB01000353 GBYB01000354 JAG70120.1 JAG70121.1 OWR52018.1 NWSH01005088 PCG64446.1 EZ407241 ACX53798.1 JN033703 AEW46856.1 NNAY01003257 OXU19723.1 KPI96444.1 KK853419 KDR07716.1 KP237872 AKD01725.1 AGBW02007651 OWR54929.1 GL888207 EGI65077.1 GBYB01002216 JAG71983.1 BT127803 AEE62765.1 SOQ53637.1 LJIJ01002106 ODM90218.1 AB701383 JN033702 AEW46855.1 KU218649 ALX00029.1 KQ414726 KOC62648.1 AGBW02010783 OWR47810.1 AAZX01000044 GALX01003699 JAB64767.1 KQ434902 KZC11162.1 KQ977349 KYN03394.1 AXCN02001099 GEYN01000021 JAV02108.1 GL768421 EFZ11357.1 KB200718 ESP00397.1 MF687618 ATJ44544.1 KQ777390 OAD52083.1 ESP00396.1 APGK01021961 APGK01021962 KB740392 ENN80755.1 KZ288235 PBC31398.1 JR040676 AEY59237.1 KU932401 APA34037.1 ODYU01001161 SOQ36985.1 GFDG01003937 JAV14862.1 GAIX01005504 JAA87056.1 GFDG01003939 JAV14860.1 GFDG01003938 JAV14861.1 MF687580 ATJ44506.1 AXCM01001195
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9JJU8
H9JJU9
A0A0H5AQ19
B8ZX07
A0A291P0V2
A0A291P0V4
+ More
G9F9G1 I4DKC0 A0A0N1PGT3 A0A212ENH0 U5KCB5 A0A194PV62 A0A2A4J392 A0A212ERG2 A0A2A4J1K6 H9IVT6 I4DKQ3 A0A291P0T7 A0A2A4JMB2 A0A1L8D6L9 A0A194RJ39 A0A2A4K2R0 Q9Y020 A0A2A4IW06 A0A291P0Y7 A0A1B2G267 A0A1L8D6N3 A0A1Z2RRK0 A0A2W1BLQ6 U5KCB8 A0A2W1BC51 G9F9G0 A0A194Q090 A0A194PKB3 I4DP16 D2SNM4 A0A2H1WKT0 A0A291P0X3 A0A2H1VAA3 U5KBY4 A0A212FE35 H9IVR7 A0A194R6Z8 A0A291P0W7 A0A212EU63 H9IVS0 A0A291P0U2 V5GQ24 A0A194PSF7 A0A291P0Y9 G9F9F4 A0A291P0S0 A0A0C9QAV8 A0A212FE89 A0A2A4IYA3 D2SNV7 G9F9F8 A0A232EMX7 A0A194PUL9 A0A067QJF4 A0A0F6Q2W6 A0A212FML6 F4WLB4 A0A0C9R605 J3JX35 A0A2H1WKV4 A0A1D2MB39 H9JTG9 G9F9F7 A0A0U4B4X4 A0A0L7QVJ2 A0A212F244 K7IXG3 A0A1W4X5Y5 V5GT06 A0A154PH71 A0A195CTD6 A0A182QZ12 A0A088AGI5 A0A1L8D6N6 E9J719 V4B296 A0A291P0Z6 A0A310SCS5 V4AZ56 N6UG67 A0A2A3EI70 V9IEZ4 A0A1I9WLL7 A0A2H1V815 A0A1L8E8F0 S4P5J6 A0A1L8E7Z6 A0A1L8E836 A0A291P0V1 A0A182M9D3
G9F9G1 I4DKC0 A0A0N1PGT3 A0A212ENH0 U5KCB5 A0A194PV62 A0A2A4J392 A0A212ERG2 A0A2A4J1K6 H9IVT6 I4DKQ3 A0A291P0T7 A0A2A4JMB2 A0A1L8D6L9 A0A194RJ39 A0A2A4K2R0 Q9Y020 A0A2A4IW06 A0A291P0Y7 A0A1B2G267 A0A1L8D6N3 A0A1Z2RRK0 A0A2W1BLQ6 U5KCB8 A0A2W1BC51 G9F9G0 A0A194Q090 A0A194PKB3 I4DP16 D2SNM4 A0A2H1WKT0 A0A291P0X3 A0A2H1VAA3 U5KBY4 A0A212FE35 H9IVR7 A0A194R6Z8 A0A291P0W7 A0A212EU63 H9IVS0 A0A291P0U2 V5GQ24 A0A194PSF7 A0A291P0Y9 G9F9F4 A0A291P0S0 A0A0C9QAV8 A0A212FE89 A0A2A4IYA3 D2SNV7 G9F9F8 A0A232EMX7 A0A194PUL9 A0A067QJF4 A0A0F6Q2W6 A0A212FML6 F4WLB4 A0A0C9R605 J3JX35 A0A2H1WKV4 A0A1D2MB39 H9JTG9 G9F9F7 A0A0U4B4X4 A0A0L7QVJ2 A0A212F244 K7IXG3 A0A1W4X5Y5 V5GT06 A0A154PH71 A0A195CTD6 A0A182QZ12 A0A088AGI5 A0A1L8D6N6 E9J719 V4B296 A0A291P0Z6 A0A310SCS5 V4AZ56 N6UG67 A0A2A3EI70 V9IEZ4 A0A1I9WLL7 A0A2H1V815 A0A1L8E8F0 S4P5J6 A0A1L8E7Z6 A0A1L8E836 A0A291P0V1 A0A182M9D3
PDB
5AZ1
E-value=1.40602e-160,
Score=1452
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00790 Folate biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
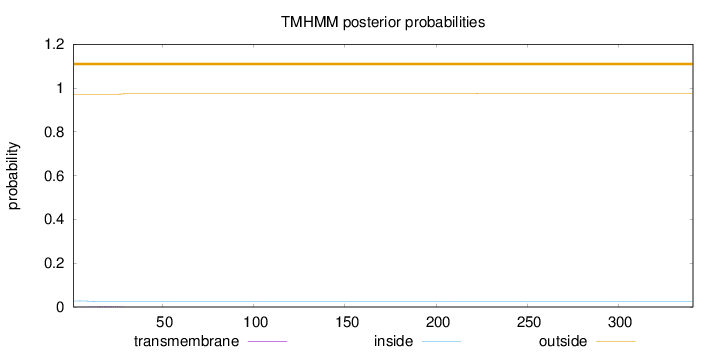
Topology
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04657
Exp number, first 60 AAs:
0.04527
Total prob of N-in:
0.02783
outside
1 - 341
Population Genetic Test Statistics
Pi
209.81011
Theta
141.221891
Tajima's D
1.177512
CLR
0.121065
CSRT
0.710514474276286
Interpretation
Uncertain
