Gene
KWMTBOMO00932
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.001
Sequence
CDS
ATGAGACGGACTTGTGAAGGCTGGGATGGTGGAGTTACCATTGGTGGAGTAAAATTGACCAACCTCCGATACGCCGACGACACCAATCTTCTCGCGGCTAATGAAAGCGAGATGGCTGTTCTCATGAGCAAGCTTGAAAAAATTAGCCTCGAACTTGGTCTTGCCATTAACCGATCCAAAACAAAAGTAATGGTAATAGACCGCATGAACAAACTGGAGCACATGGGGTCACTGCATCTTGAAACAACAGAAAGATTCATATACCTGGGCTCCATGATTGCGAATACTGGATCGTGCGAGCCTGAGATTCGAAGACGTATCGGGATAGCAAAAACTGCCATGTCTCAGCTTGATAGCATCTGGTCCGATAGAAACATCTCCATAAAAACCAAGCGTAAACTTGTAACAAGTCTTGTGTTTTCTATTTTCTTATATGGGGCGGAGACATGGACTCTGAAGAAGGCTGATCGAAGTCAGATCGACGCGTTTGAGATGTGGTGCTGGAGGAAAATGCTGCGGATTCCCTGGACGGCGCATCGTACAAACGCATCAATCCTGGAAGAACTAGGCGTCTCCTTAAGACTCTCCACCATTTGCCTACGTAGGATTTTAGAGTTTTTCGGTCACGTTGCGAGAAAAAAGGGTGACAATCTCGAAAGACTTCTCATCACCGGCAAAGTGGAAGGAAAGAGGCCTAGGGGGCGTAGTCCCATGCGCTGGTCCGATCAGATCAAGACGACTTTGGAATCCAGCATCAATGTCGCTATCCACTGCGCGGAGGACAGAAGTGAATGGAAGAACATAGTCCTGACAAAAGTGCTCAGTAAAGGTCACGACCCTCAGAACTGA
Protein
MRRTCEGWDGGVTIGGVKLTNLRYADDTNLLAANESEMAVLMSKLEKISLELGLAINRSKTKVMVIDRMNKLEHMGSLHLETTERFIYLGSMIANTGSCEPEIRRRIGIAKTAMSQLDSIWSDRNISIKTKRKLVTSLVFSIFLYGAETWTLKKADRSQIDAFEMWCWRKMLRIPWTAHRTNASILEELGVSLRLSTICLRRILEFFGHVARKKGDNLERLLITGKVEGKRPRGRSPMRWSDQIKTTLESSINVAIHCAEDRSEWKNIVLTKVLSKGHDPQN
Summary
Uniprot
D7F163
A0A2H1VU13
D7F168
D7F172
D7F161
A0A2W1BGD9
+ More
D7F162 D7F167 D7F175 D5LB39 A0A2G8JE23 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 A0A1J5WD64 H2YWZ5 A0A2H6KKM9 A0A061BKJ9 W4YPU3 A0A061BKT1 A0A0B7BUR9 O97916 A0A023G885 H3B2Y3 A0A2A4J795 A0A0B7BVD3 A0A146L0Z1 W5PUC3 A0A2G8JVA1 A0A2G8K183 X1D4L4 W4XFH1 W5NGM3 W4YNG7 W4Z5A1 A0A2V9X8L4 D5LB38 H2YWZ6 W5MYT3 W5N9W4 A0A2S2PJ33 W5MW50 A0A0J7KZE8 A0A2S2PMS6 A0A0A9XXL9 A0A0B7BSZ9 A0A2S2QAF2 W4Z133 A0A0B7BVD9 A0A0J7KRQ5 W5MYU0 A0A2B8A3L2 W5NIE2 A0A2S2PZ46 W4Y921 A0A2G8JCI6 A0A0A9WMP5 A0A0P4W0B7 A0A0A9XCF5 A0A0B7BT80 A0A0A9WVQ5 W4XIJ3 W4YB77 A0A146M2R9 A0A2S2QE77 A0A0B7BSB6 A0A146LUY8 A0A2A4JXP9 W5MUQ0 X1XQS0 A0A2G8JCM7 H3J6U9 H3AY06 J9KDM4 W4XHV2 J9LZK0 A0A2S2Q0B1 A0A076N1B6 A0A2J7R4L3 A0A3B3HVR8 A0A2J7RFE8 W5MW18
D7F162 D7F167 D7F175 D5LB39 A0A2G8JE23 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 A0A1J5WD64 H2YWZ5 A0A2H6KKM9 A0A061BKJ9 W4YPU3 A0A061BKT1 A0A0B7BUR9 O97916 A0A023G885 H3B2Y3 A0A2A4J795 A0A0B7BVD3 A0A146L0Z1 W5PUC3 A0A2G8JVA1 A0A2G8K183 X1D4L4 W4XFH1 W5NGM3 W4YNG7 W4Z5A1 A0A2V9X8L4 D5LB38 H2YWZ6 W5MYT3 W5N9W4 A0A2S2PJ33 W5MW50 A0A0J7KZE8 A0A2S2PMS6 A0A0A9XXL9 A0A0B7BSZ9 A0A2S2QAF2 W4Z133 A0A0B7BVD9 A0A0J7KRQ5 W5MYU0 A0A2B8A3L2 W5NIE2 A0A2S2PZ46 W4Y921 A0A2G8JCI6 A0A0A9WMP5 A0A0P4W0B7 A0A0A9XCF5 A0A0B7BT80 A0A0A9WVQ5 W4XIJ3 W4YB77 A0A146M2R9 A0A2S2QE77 A0A0B7BSB6 A0A146LUY8 A0A2A4JXP9 W5MUQ0 X1XQS0 A0A2G8JCM7 H3J6U9 H3AY06 J9KDM4 W4XHV2 J9LZK0 A0A2S2Q0B1 A0A076N1B6 A0A2J7R4L3 A0A3B3HVR8 A0A2J7RFE8 W5MW18
Pubmed
EMBL
FJ265548
ADI61816.1
ODYU01004401
SOQ44236.1
FJ265553
ADI61821.1
+ More
FJ265557 ADI61825.1 FJ265546 ADI61814.1 KZ150203 PZC72267.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 MRZV01002322 PIK33998.1 MRZV01000196 PIK55924.1 MRZV01001207 PIK39730.1 AHAT01022938 MKPU01000878 OIR55924.1 BDSA01000081 GBE63537.1 LK055282 CDR71995.1 AAGJ04055194 LK054988 CDR71535.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 AJ132772 CAA10770.1 GBBM01006298 JAC29120.1 AFYH01119920 NWSH01002736 PCG67606.1 HACG01049245 CEK96110.1 GDHC01017214 JAQ01415.1 AMGL01091659 AMGL01091660 AMGL01091661 AMGL01091662 AMGL01091663 AMGL01091664 AMGL01091665 AMGL01091666 AMGL01091667 AMGL01091668 MRZV01001216 PIK39650.1 MRZV01000994 PIK41715.1 BART01031562 GAH15691.1 AAGJ04075245 AHAT01008485 AAGJ04167631 AAGJ04128321 QIAP01000240 PYX61602.1 GU815089 ADF18552.1 AHAT01017643 AHAT01022314 GGMR01016842 MBY29461.1 AHAT01013219 LBMM01001656 KMQ95932.1 GGMR01018096 MBY30715.1 GBHO01020011 GDHC01013600 JAG23593.1 JAQ05029.1 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 GGMS01004979 MBY74182.1 AAGJ04171899 HACG01049255 CEK96120.1 LBMM01003995 KMQ92919.1 PDHN01000302 PGH39547.1 AHAT01009145 GGMS01001437 MBY70640.1 AAGJ04161766 MRZV01002544 PIK33458.1 GBHO01034555 JAG09049.1 GDRN01098129 JAI59014.1 GBHO01028834 JAG14770.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 GBHO01033031 GBHO01015473 GBHO01015472 GBHO01015471 GBHO01015469 GBHO01013134 GBHO01013133 GBHO01013129 GBHO01013128 JAG10573.1 JAG28131.1 JAG28132.1 JAG28133.1 JAG28135.1 JAG30470.1 JAG30471.1 JAG30475.1 JAG30476.1 AAGJ04122993 AAGJ04093867 GDHC01004555 JAQ14074.1 GGMS01006790 MBY75993.1 HACG01049249 CEK96114.1 GDHC01007098 GDHC01001590 JAQ11531.1 JAQ17039.1 NWSH01000469 PCG76200.1 AHAT01016497 ABLF02038495 MRZV01002526 PIK33493.1 AAGJ04118689 AAGJ04161747 AFYH01053121 ABLF02046942 ABLF02059971 AAGJ04165736 ABLF02042206 GGMS01001369 MBY70572.1 KF881087 AIJ27486.1 NEVH01007405 PNF35773.1 NEVH01004413 PNF39563.1 AHAT01013157 AHAT01013158
FJ265557 ADI61825.1 FJ265546 ADI61814.1 KZ150203 PZC72267.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 MRZV01002322 PIK33998.1 MRZV01000196 PIK55924.1 MRZV01001207 PIK39730.1 AHAT01022938 MKPU01000878 OIR55924.1 BDSA01000081 GBE63537.1 LK055282 CDR71995.1 AAGJ04055194 LK054988 CDR71535.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 AJ132772 CAA10770.1 GBBM01006298 JAC29120.1 AFYH01119920 NWSH01002736 PCG67606.1 HACG01049245 CEK96110.1 GDHC01017214 JAQ01415.1 AMGL01091659 AMGL01091660 AMGL01091661 AMGL01091662 AMGL01091663 AMGL01091664 AMGL01091665 AMGL01091666 AMGL01091667 AMGL01091668 MRZV01001216 PIK39650.1 MRZV01000994 PIK41715.1 BART01031562 GAH15691.1 AAGJ04075245 AHAT01008485 AAGJ04167631 AAGJ04128321 QIAP01000240 PYX61602.1 GU815089 ADF18552.1 AHAT01017643 AHAT01022314 GGMR01016842 MBY29461.1 AHAT01013219 LBMM01001656 KMQ95932.1 GGMR01018096 MBY30715.1 GBHO01020011 GDHC01013600 JAG23593.1 JAQ05029.1 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 GGMS01004979 MBY74182.1 AAGJ04171899 HACG01049255 CEK96120.1 LBMM01003995 KMQ92919.1 PDHN01000302 PGH39547.1 AHAT01009145 GGMS01001437 MBY70640.1 AAGJ04161766 MRZV01002544 PIK33458.1 GBHO01034555 JAG09049.1 GDRN01098129 JAI59014.1 GBHO01028834 JAG14770.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 GBHO01033031 GBHO01015473 GBHO01015472 GBHO01015471 GBHO01015469 GBHO01013134 GBHO01013133 GBHO01013129 GBHO01013128 JAG10573.1 JAG28131.1 JAG28132.1 JAG28133.1 JAG28135.1 JAG30470.1 JAG30471.1 JAG30475.1 JAG30476.1 AAGJ04122993 AAGJ04093867 GDHC01004555 JAQ14074.1 GGMS01006790 MBY75993.1 HACG01049249 CEK96114.1 GDHC01007098 GDHC01001590 JAQ11531.1 JAQ17039.1 NWSH01000469 PCG76200.1 AHAT01016497 ABLF02038495 MRZV01002526 PIK33493.1 AAGJ04118689 AAGJ04161747 AFYH01053121 ABLF02046942 ABLF02059971 AAGJ04165736 ABLF02042206 GGMS01001369 MBY70572.1 KF881087 AIJ27486.1 NEVH01007405 PNF35773.1 NEVH01004413 PNF39563.1 AHAT01013157 AHAT01013158
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D7F163
A0A2H1VU13
D7F168
D7F172
D7F161
A0A2W1BGD9
+ More
D7F162 D7F167 D7F175 D5LB39 A0A2G8JE23 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 A0A1J5WD64 H2YWZ5 A0A2H6KKM9 A0A061BKJ9 W4YPU3 A0A061BKT1 A0A0B7BUR9 O97916 A0A023G885 H3B2Y3 A0A2A4J795 A0A0B7BVD3 A0A146L0Z1 W5PUC3 A0A2G8JVA1 A0A2G8K183 X1D4L4 W4XFH1 W5NGM3 W4YNG7 W4Z5A1 A0A2V9X8L4 D5LB38 H2YWZ6 W5MYT3 W5N9W4 A0A2S2PJ33 W5MW50 A0A0J7KZE8 A0A2S2PMS6 A0A0A9XXL9 A0A0B7BSZ9 A0A2S2QAF2 W4Z133 A0A0B7BVD9 A0A0J7KRQ5 W5MYU0 A0A2B8A3L2 W5NIE2 A0A2S2PZ46 W4Y921 A0A2G8JCI6 A0A0A9WMP5 A0A0P4W0B7 A0A0A9XCF5 A0A0B7BT80 A0A0A9WVQ5 W4XIJ3 W4YB77 A0A146M2R9 A0A2S2QE77 A0A0B7BSB6 A0A146LUY8 A0A2A4JXP9 W5MUQ0 X1XQS0 A0A2G8JCM7 H3J6U9 H3AY06 J9KDM4 W4XHV2 J9LZK0 A0A2S2Q0B1 A0A076N1B6 A0A2J7R4L3 A0A3B3HVR8 A0A2J7RFE8 W5MW18
D7F162 D7F167 D7F175 D5LB39 A0A2G8JE23 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 A0A1J5WD64 H2YWZ5 A0A2H6KKM9 A0A061BKJ9 W4YPU3 A0A061BKT1 A0A0B7BUR9 O97916 A0A023G885 H3B2Y3 A0A2A4J795 A0A0B7BVD3 A0A146L0Z1 W5PUC3 A0A2G8JVA1 A0A2G8K183 X1D4L4 W4XFH1 W5NGM3 W4YNG7 W4Z5A1 A0A2V9X8L4 D5LB38 H2YWZ6 W5MYT3 W5N9W4 A0A2S2PJ33 W5MW50 A0A0J7KZE8 A0A2S2PMS6 A0A0A9XXL9 A0A0B7BSZ9 A0A2S2QAF2 W4Z133 A0A0B7BVD9 A0A0J7KRQ5 W5MYU0 A0A2B8A3L2 W5NIE2 A0A2S2PZ46 W4Y921 A0A2G8JCI6 A0A0A9WMP5 A0A0P4W0B7 A0A0A9XCF5 A0A0B7BT80 A0A0A9WVQ5 W4XIJ3 W4YB77 A0A146M2R9 A0A2S2QE77 A0A0B7BSB6 A0A146LUY8 A0A2A4JXP9 W5MUQ0 X1XQS0 A0A2G8JCM7 H3J6U9 H3AY06 J9KDM4 W4XHV2 J9LZK0 A0A2S2Q0B1 A0A076N1B6 A0A2J7R4L3 A0A3B3HVR8 A0A2J7RFE8 W5MW18
Ontologies
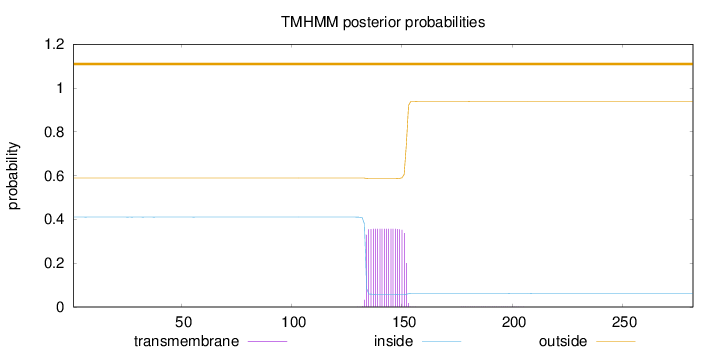
Topology
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.63721
Exp number, first 60 AAs:
0.00245
Total prob of N-in:
0.41075
outside
1 - 282
Population Genetic Test Statistics
Pi
319.699137
Theta
172.777039
Tajima's D
2.659689
CLR
12.92772
CSRT
0.958002099895005
Interpretation
Uncertain
