Gene
KWMTBOMO00931
Pre Gene Modal
BGIBMGA009797
Annotation
segmentation_polarity_homeobox_protein_engrailed_[Bombyx_mori]
Full name
Segmentation polarity homeobox protein engrailed
+ More
Homeobox protein engrailed-like
Homeobox protein engrailed-like
Location in the cell
Nuclear Reliability : 4.28
Sequence
CDS
ATGGCCTTCGAGGACCGCTGCAGCCCTAGCCAGGCTAACAGTCCGGGACCGGTGACGGGGCGAGTCCCGGCGCCGCACGCCGAGACCCTCGCATACAGCCCGCAGAGCCAGTACACTTGCACCACAATAGAATCGAAGTACGAACGAGGCTCTCCGAACATGACAATTGTGAAGGTGCAGCCTGACTCCCCGCCTCCCAGCCCGGGACGCGGTCAGAACGAGATGGAATACCAGGACTACTATCGCCCTGAAACGCCCGACGTAAAGCCACACTTCAGCCGGGAGGAGCAGAGGTTTGAACTGGACAGATCGAGGGGGCAGCGGCTGCAACCCACCACGCCGGTCGCCTTCTCCATAAACAACATCCTGCACCCTGAGTTCGGCTTGAACGCCATCAGGAAAACGAACAAAATCGAAGGTCCCAAGCCCATTGGACCGAACCACAGTATCCTCTATAAGCCTTACGATTTATCCAAACCGGACTTATCGAAATACGGCTTTGATTATTTGAAGAGTAAGGAAACGAGTGATTGCAACGCTTTGCCGCCTTTAGGAGGGTTGAGGGAGACGGTATCGCAGATCGGCGAACGCTTGTCCAGAGACAGGGAGCCTCCGAAGAGTCTGGAGCAGCAGAAGAGGCCCGACTCCGCGAGTTCCATCGTCTCGTCGACGTCAAGCGGCGCGGTTTCCACCTGCGGAAGCTCGGACGCGAGCTCCATCCAGTCCCAGAGCAACCCTGGCCAGCTGTGGCCAGCCTGGGTATACTGCACCAGATACAGCGACCGACCTAGTTCCGGTGAGTGCAAATTTTAA
Protein
MAFEDRCSPSQANSPGPVTGRVPAPHAETLAYSPQSQYTCTTIESKYERGSPNMTIVKVQPDSPPPSPGRGQNEMEYQDYYRPETPDVKPHFSREEQRFELDRSRGQRLQPTTPVAFSINNILHPEFGLNAIRKTNKIEGPKPIGPNHSILYKPYDLSKPDLSKYGFDYLKSKETSDCNALPPLGGLRETVSQIGERLSRDREPPKSLEQQKRPDSASSIVSSTSSGAVSTCGSSDASSIQSQSNPGQLWPAWVYCTRYSDRPSSGECKF
Summary
Similarity
Belongs to the engrailed homeobox family.
Belongs to the Engrailed homeobox family.
Belongs to the Engrailed homeobox family.
Keywords
Complete proteome
Developmental protein
DNA-binding
Homeobox
Nucleus
Reference proteome
Segmentation polarity protein
Feature
chain Segmentation polarity homeobox protein engrailed
Uniprot
H9JJU6
P27609
A0A2H1V3I6
A0A2A4IWU5
A0A0N0PAN3
A0A0N0PDN7
+ More
B9WPT6 A0A024CBQ7 A0A024CAM5 A0A024CCY6 A0A024CB14 A0A024CB83 A0A024CBP7 A0A024CBR5 A0A024CB77 A0A024CCW5 A0A024CBR1 A0A024CAZ8 A0A024CB08 A0A024CB21 A0A024CCX5 A0A024CCV8 A0A024CCX0 A0A024CAP9 A0A024CBA4 A0A024CAZ2 A0A0L7L0G9 A0A024CAL9 A0A024CB72 A0A024CBP1 A0A194PV56 A0A024CAN9 A0A024CCY0 A0A023UN14 A0A023UMY3 A0A024CBQ2 A0A023ULZ6 A0A023UN10 A0A023UMI7 A0A023UMG8 A0A023UL75 A0A023UMJ1 A0A023ULZ0 A0A023ULA4 A0A023UKN1 A0A023UMY9 A0A023ULW2 A0A023UKQ6 A0A023ULV8 A0A023UKM0 A0A023UL67 A0A023UMG1 A0A023UKR2 A0A023UMH2 A0A023UL80 A0A024CB92 A0A024CCZ7 A0A024CAP4 I4DQU6
B9WPT6 A0A024CBQ7 A0A024CAM5 A0A024CCY6 A0A024CB14 A0A024CB83 A0A024CBP7 A0A024CBR5 A0A024CB77 A0A024CCW5 A0A024CBR1 A0A024CAZ8 A0A024CB08 A0A024CB21 A0A024CCX5 A0A024CCV8 A0A024CCX0 A0A024CAP9 A0A024CBA4 A0A024CAZ2 A0A0L7L0G9 A0A024CAL9 A0A024CB72 A0A024CBP1 A0A194PV56 A0A024CAN9 A0A024CCY0 A0A023UN14 A0A023UMY3 A0A024CBQ2 A0A023ULZ6 A0A023UN10 A0A023UMI7 A0A023UMG8 A0A023UL75 A0A023UMJ1 A0A023ULZ0 A0A023ULA4 A0A023UKN1 A0A023UMY9 A0A023ULW2 A0A023UKQ6 A0A023ULV8 A0A023UKM0 A0A023UL67 A0A023UMG1 A0A023UKR2 A0A023UMH2 A0A023UL80 A0A024CB92 A0A024CCZ7 A0A024CAP4 I4DQU6
EMBL
BABH01017824
M64335
ODYU01000344
SOQ34932.1
NWSH01006213
PCG63590.1
+ More
KQ458706 KPJ05484.1 KQ460280 KPJ16309.1 FM995623 CAX36786.1 KJ507636 AHZ31555.1 KJ507624 AHZ31543.1 KJ507643 AHZ31562.1 KJ507640 AHZ31559.1 KJ507632 AHZ31551.1 KJ507626 KJ507629 AHZ31545.1 KJ507646 AHZ31565.1 KJ507627 KJ507630 AHZ31546.1 KJ507623 AHZ31542.1 KJ507641 AHZ31560.1 KJ507625 AHZ31544.1 KJ507635 KJ507637 AHZ31554.1 KJ507645 AHZ31564.1 KJ507633 KJ507634 AHZ31552.1 KJ507618 AHZ31537.1 KJ507628 AHZ31547.1 KJ507648 KJ507649 KJ507650 KJ507651 AHZ31568.1 KJ507652 AHZ31571.1 KJ507620 AHZ31539.1 JTDY01003936 KOB68811.1 KJ507619 AHZ31538.1 KJ507622 AHZ31541.1 KJ507621 AHZ31540.1 KQ459593 KPI96639.1 KJ507639 AHZ31558.1 KJ507638 AHZ31557.1 KJ458803 KJ458827 AHY04072.1 KJ458797 AHY04042.1 KJ507631 AHZ31550.1 KJ458828 AHY04073.1 KJ458822 AHY04067.1 KJ458821 AHY04066.1 KJ458801 AHY04046.1 KJ458799 KJ458800 AHY04045.1 KJ458826 AHY04071.1 KJ458823 AHY04068.1 KJ458825 AHY04070.1 KJ458804 AHY04049.1 KJ458802 AHY04047.1 KJ458798 AHY04043.1 KJ458824 AHY04069.1 KJ458793 AHY04038.1 KJ458794 AHY04039.1 KJ458795 AHY04040.1 KJ458796 AHY04041.1 KJ458829 AHY04074.1 KJ458806 AHY04051.1 KJ458805 AHY04050.1 KJ507642 AHZ31561.1 KJ507653 AHZ31572.1 KJ507644 AHZ31563.1 AK404792 BAM20286.1
KQ458706 KPJ05484.1 KQ460280 KPJ16309.1 FM995623 CAX36786.1 KJ507636 AHZ31555.1 KJ507624 AHZ31543.1 KJ507643 AHZ31562.1 KJ507640 AHZ31559.1 KJ507632 AHZ31551.1 KJ507626 KJ507629 AHZ31545.1 KJ507646 AHZ31565.1 KJ507627 KJ507630 AHZ31546.1 KJ507623 AHZ31542.1 KJ507641 AHZ31560.1 KJ507625 AHZ31544.1 KJ507635 KJ507637 AHZ31554.1 KJ507645 AHZ31564.1 KJ507633 KJ507634 AHZ31552.1 KJ507618 AHZ31537.1 KJ507628 AHZ31547.1 KJ507648 KJ507649 KJ507650 KJ507651 AHZ31568.1 KJ507652 AHZ31571.1 KJ507620 AHZ31539.1 JTDY01003936 KOB68811.1 KJ507619 AHZ31538.1 KJ507622 AHZ31541.1 KJ507621 AHZ31540.1 KQ459593 KPI96639.1 KJ507639 AHZ31558.1 KJ507638 AHZ31557.1 KJ458803 KJ458827 AHY04072.1 KJ458797 AHY04042.1 KJ507631 AHZ31550.1 KJ458828 AHY04073.1 KJ458822 AHY04067.1 KJ458821 AHY04066.1 KJ458801 AHY04046.1 KJ458799 KJ458800 AHY04045.1 KJ458826 AHY04071.1 KJ458823 AHY04068.1 KJ458825 AHY04070.1 KJ458804 AHY04049.1 KJ458802 AHY04047.1 KJ458798 AHY04043.1 KJ458824 AHY04069.1 KJ458793 AHY04038.1 KJ458794 AHY04039.1 KJ458795 AHY04040.1 KJ458796 AHY04041.1 KJ458829 AHY04074.1 KJ458806 AHY04051.1 KJ458805 AHY04050.1 KJ507642 AHZ31561.1 KJ507653 AHZ31572.1 KJ507644 AHZ31563.1 AK404792 BAM20286.1
Proteomes
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
H9JJU6
P27609
A0A2H1V3I6
A0A2A4IWU5
A0A0N0PAN3
A0A0N0PDN7
+ More
B9WPT6 A0A024CBQ7 A0A024CAM5 A0A024CCY6 A0A024CB14 A0A024CB83 A0A024CBP7 A0A024CBR5 A0A024CB77 A0A024CCW5 A0A024CBR1 A0A024CAZ8 A0A024CB08 A0A024CB21 A0A024CCX5 A0A024CCV8 A0A024CCX0 A0A024CAP9 A0A024CBA4 A0A024CAZ2 A0A0L7L0G9 A0A024CAL9 A0A024CB72 A0A024CBP1 A0A194PV56 A0A024CAN9 A0A024CCY0 A0A023UN14 A0A023UMY3 A0A024CBQ2 A0A023ULZ6 A0A023UN10 A0A023UMI7 A0A023UMG8 A0A023UL75 A0A023UMJ1 A0A023ULZ0 A0A023ULA4 A0A023UKN1 A0A023UMY9 A0A023ULW2 A0A023UKQ6 A0A023ULV8 A0A023UKM0 A0A023UL67 A0A023UMG1 A0A023UKR2 A0A023UMH2 A0A023UL80 A0A024CB92 A0A024CCZ7 A0A024CAP4 I4DQU6
B9WPT6 A0A024CBQ7 A0A024CAM5 A0A024CCY6 A0A024CB14 A0A024CB83 A0A024CBP7 A0A024CBR5 A0A024CB77 A0A024CCW5 A0A024CBR1 A0A024CAZ8 A0A024CB08 A0A024CB21 A0A024CCX5 A0A024CCV8 A0A024CCX0 A0A024CAP9 A0A024CBA4 A0A024CAZ2 A0A0L7L0G9 A0A024CAL9 A0A024CB72 A0A024CBP1 A0A194PV56 A0A024CAN9 A0A024CCY0 A0A023UN14 A0A023UMY3 A0A024CBQ2 A0A023ULZ6 A0A023UN10 A0A023UMI7 A0A023UMG8 A0A023UL75 A0A023UMJ1 A0A023ULZ0 A0A023ULA4 A0A023UKN1 A0A023UMY9 A0A023ULW2 A0A023UKQ6 A0A023ULV8 A0A023UKM0 A0A023UL67 A0A023UMG1 A0A023UKR2 A0A023UMH2 A0A023UL80 A0A024CB92 A0A024CCZ7 A0A024CAP4 I4DQU6
Ontologies
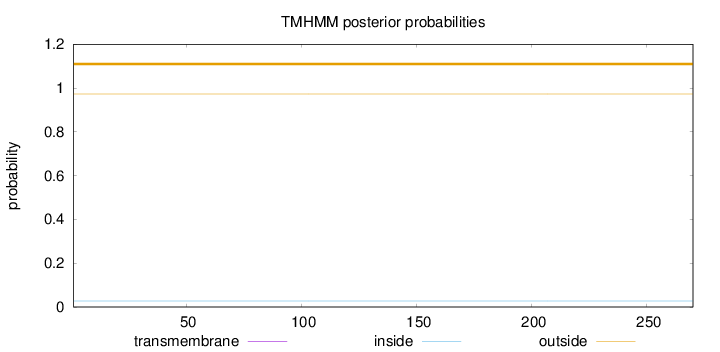
Topology
Subcellular location
Nucleus
Length:
270
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00084
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02740
outside
1 - 270
Population Genetic Test Statistics
Pi
299.885584
Theta
227.681724
Tajima's D
0.910939
CLR
0.22427
CSRT
0.629618519074046
Interpretation
Uncertain
