Gene
KWMTBOMO00929
Annotation
reverse_transcriptase_[Lasius_niger]
Full name
Probable RNA-directed DNA polymerase from transposon X-element
Alternative Name
Reverse transcriptase
Location in the cell
Mitochondrial Reliability : 1.97 Nuclear Reliability : 2.317
Sequence
CDS
ATGCACCCAGATGGCTCCGCCCACGGAGGAAGTGCAATTATAATCAGAAGTAACATCAAGCACTATGAAGGATCTCATTACTGTACTGATATAATACAAGCAACAAACATCGTTGTTCAAGACTGGGTGGGACCCATCACCATATCGGCACTTTACAGCCCTCCAAAACATAACATTAAGAATGAAGATTATGAAAAGTTTTTTAATTCCTTAGGACATAGATTTATGGCTGGTGGAGATTATAATGCTAAGCATACAAGCTGGGGGTCTCGAACTGTGACAACGAAGGGAAGACAGTTACTTTCTTGCATTGAACGACTAAGATTAAATGTTTGTTCAACTGGCGAGCCAACATATTGGCCGAGTGACAAAAGGAAGGTACCAGACTTGTTAGATTTTTGTATCACAAAAGGCATCAATAATGCATGTATAGAGTGTACATCCAGTTTTGATCTCTCATCTGATCACTCCCCAGTCATAATAACGCTCAGTCGAAATATAGCCAGGAAAATGAAAACATGTAGACTGCATTCTTGCAGGACCGATTGGGATTACTTTCGTGTATTGGTATCTGGCTCTTTGGACATCAATCTACCTCTAAAAACAGAGGATGACATTGCGGATGGTGTTGAACATTTTAACTTTGTTGTACAACATTCTGCTTGGCATGCGACACCCGCCCCACCAATTGTAAAACCATCGCAATGTTCTCTTTCAATCAAAACGTTGTTATCAGAAAAAAGGCGATTAAGAAAACGATGGCAAACTACGAGGCATCCTGATGACAAAAAAATATTTAATTACAGTATTACAAAATTGAAAATACATTTGGCTCGTGAGAGAAACGAAAAAACAAATACATATATGCAAGAGCTGGATGCCACATCGAAAACAGACTATTCGCTATGGAAGGCTACCAAATACCTTAAACGTCCTATAAACCACCAACCTCCTCTACGGAAATCTGATGGAACTTGGGCCACTAGCGACCTAGACAAAGCTAAGGCCTTTGCTGAACACCTGGATAATGTATTTACACTAAATGCGATGGATAATTCGGCTATGCCTGCCAATGAATCGGAATTGAATGAGCATGTTATTGAGGTTCCCTTCAAAAAAATTCGTAAATCCGAAACAAGATCTGTCATAAAGAATCTTGACCCTAATAAATCACCAGGATATGATCTAATAACAGCAACTATTTTAAAAGAGTTACCTCAAGAGGGAATTACTTTCTTGACCCAATTATACAATGCTATTTTCAGACTTAAGTTTGTGCCTCACTTATGGAAGGTAGCTCAAATTAAGATGATACCTAAACCTGGAAAAAGTGGCGAGGACGTGAAATCTTATCGTCCTATCAGTTTGCTCCCTGTACCGTCCAAGGTCATGGAGATTCTGTTTCTCGCAAGATTGAATCCTATTATTGAAACGAAAAAACTAATCCCAGATCATCAGTTTGGCTTTAGATGGAAACATAGTACAATCGAACAAATTCACAGACTTGTTGAATATATTAATAGAGCTTTTGAGGGAAAAAAATACTGCACGGCTGCCTTCCTAGATATATCTCAGGCCTTCGACCGAGTTTGGCATGAAGGACTCTTACTCAAAATTAAAAAGAAGATCCCTTCCAACTTCTATTTTTTCATTGAATCTTATTTGAGTAGCCGATATTATTTTGTTAAACACGGCGATGAAATATCGCCTCTCCATTCAATTAATGCCGGTGTTCCCCAAGGCAGTGTCATTGGGCCCACTTTATATCTATTGTATACAGCTGATTTGCCCGAAACTGATGGTACGATAATGGGAACTTTTGCTGATGACACCGCAATAATGACCATGGACGATGATCCAAACGAAGCTTCTTATAAACTACAGAACAGCCTTAATAATATCAACACTTGGCTAAAAAAATGGCGTATCAAAGCGAATGAATCTAAGTCTGTACAAGTTACTTTCACCACTCGAAGACAAACTTGTCCCGCAGTCACCTTGAATAATATAGAGATCCCGCAATCTGAAAATGCAAAGTACTTAGGGATGTACCTGGATAGGAGACTGACTTGGAGAAAACATATATTCACCAAGCGTCTTGCGGTGGGTCAGCAAATACGTAAATTGTATTGGCTTCTTAATAGAAAGTCTCAACTGTCAGTTGAAAACAAGCTTTTACTATACAAATCTGTTATTAAACCCATTTGGTCATATGGCATCCAATTGTGGGGTACTGCGTCAAACTCAAATATTGAAATACTTCAAAGATTTCAATCAAAAACGCTCAGGATGATAGTGAACGCACCATGGTATGTTACAAATGAAAGAATCCATTATGATCTTGCCATAAGAAAAGTCAAACATGAAGTAATCCATCGAGCTAAAGCATATTGTGTAAGATTGGAAGCCCATCCGAATAACCTTGCTAAGCATTTAATGGACAACCTAATGACTACTAGAAGACTCAGAAGAAGAGTGCCTCAAGATCTACTAGATCTACTGTAG
Protein
MHPDGSAHGGSAIIIRSNIKHYEGSHYCTDIIQATNIVVQDWVGPITISALYSPPKHNIKNEDYEKFFNSLGHRFMAGGDYNAKHTSWGSRTVTTKGRQLLSCIERLRLNVCSTGEPTYWPSDKRKVPDLLDFCITKGINNACIECTSSFDLSSDHSPVIITLSRNIARKMKTCRLHSCRTDWDYFRVLVSGSLDINLPLKTEDDIADGVEHFNFVVQHSAWHATPAPPIVKPSQCSLSIKTLLSEKRRLRKRWQTTRHPDDKKIFNYSITKLKIHLARERNEKTNTYMQELDATSKTDYSLWKATKYLKRPINHQPPLRKSDGTWATSDLDKAKAFAEHLDNVFTLNAMDNSAMPANESELNEHVIEVPFKKIRKSETRSVIKNLDPNKSPGYDLITATILKELPQEGITFLTQLYNAIFRLKFVPHLWKVAQIKMIPKPGKSGEDVKSYRPISLLPVPSKVMEILFLARLNPIIETKKLIPDHQFGFRWKHSTIEQIHRLVEYINRAFEGKKYCTAAFLDISQAFDRVWHEGLLLKIKKKIPSNFYFFIESYLSSRYYFVKHGDEISPLHSINAGVPQGSVIGPTLYLLYTADLPETDGTIMGTFADDTAIMTMDDDPNEASYKLQNSLNNINTWLKKWRIKANESKSVQVTFTTRRQTCPAVTLNNIEIPQSENAKYLGMYLDRRLTWRKHIFTKRLAVGQQIRKLYWLLNRKSQLSVENKLLLYKSVIKPIWSYGIQLWGTASNSNIEILQRFQSKTLRMIVNAPWYVTNERIHYDLAIRKVKHEVIHRAKAYCVRLEAHPNNLAKHLMDNLMTTRRLRRRVPQDLLDLL
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
Mg(2+)
Mn(2+)
Mn(2+)
Miscellaneous
At least 30 copies of X-element are present in the genome and some elements are polymorphic between different strains.
Keywords
Nucleotidyltransferase
RNA-directed DNA polymerase
Transferase
Transposable element
Feature
chain Probable RNA-directed DNA polymerase from transposon X-element
Uniprot
A0A0J7MYU2
A0A2J7R4U3
A0A2J7Q9Z1
A0A2J7QES2
A0A2J7PSM7
A0A2J7R7D0
+ More
A0A2J7R098 A0A2J7RQ52 A0A2J7PMN4 A0A2J7PVX3 A0A2J7QB68 A0A2A4K918 A0A2A4IUM2 A0A2J7Q0S7 A0A0J7K4E0 A0A2J7PDM0 J9LYS0 A0A1Y1KFR3 A0A1Y1LKN6 A0A0A9YU04 A0A0A9YJ00 A0A2J7RD70 A0A0J7K811 A0A142LX26 A0A1B6EGD8 A0A142LX53 A0A2J7PVE7 A0A034VFD2 Q6R811 Q04135 A0A0J7K7J8 A0A142LX49 Q961V7 A0A0M4EWX7 Q24335 A0A224X6T4 A0A352X8G4 A0A1L8DIZ6 A0A0J7NDW4 A0A069DX78 A0A224XHW8 A0A0V0G5B6 A0A2S2NGV8 A0A0A9YSX4 A0A2S2N814 A0A224X746 A0A2J7R8R8 A0A224XJI0 A0A224X6R6 A0A023F0I3 A0A2S2N6Q7 A0A224X724 X1WUG2 A0A2S2PJG2 A0A2S2P698 A0A2S2NH02 A0A224X6U5 A0A224XIB0 A0A224X6W2 A0A224X6S1 A0A2S2P6S3 A0A224XI99 A0A2S2NPJ7 A0A2S2R9D4 A0A2H8TKI9 A0A2S2PHL7 A0A2H8TF37 A0A0J7K3N0 A0A2S2P6C4 A0A023F069 A0A2S2PUS3 A0A2S2QDN9 A0A2S2P7B2 A0A2S2P3X8 A0A1Y1MT28 A0A146KRW8 J9KE07 J9M0H4 A0A2S2NKC3 A0A146KV64 A0A224XK92 A0A1B6C2V8 J9KSK5 A0A2S2QEE0 Q9NBX4 A0A2S2PA10 X1XLN5 A0A2S2NPG3 J9KWJ0 A0A224X7Z3 J9M4H5 A0A2S2N9P4 X1WVQ4 J9LMA3 A0A2S2PBB4 J9KLB1 A0A2S2Q2R8
A0A2J7R098 A0A2J7RQ52 A0A2J7PMN4 A0A2J7PVX3 A0A2J7QB68 A0A2A4K918 A0A2A4IUM2 A0A2J7Q0S7 A0A0J7K4E0 A0A2J7PDM0 J9LYS0 A0A1Y1KFR3 A0A1Y1LKN6 A0A0A9YU04 A0A0A9YJ00 A0A2J7RD70 A0A0J7K811 A0A142LX26 A0A1B6EGD8 A0A142LX53 A0A2J7PVE7 A0A034VFD2 Q6R811 Q04135 A0A0J7K7J8 A0A142LX49 Q961V7 A0A0M4EWX7 Q24335 A0A224X6T4 A0A352X8G4 A0A1L8DIZ6 A0A0J7NDW4 A0A069DX78 A0A224XHW8 A0A0V0G5B6 A0A2S2NGV8 A0A0A9YSX4 A0A2S2N814 A0A224X746 A0A2J7R8R8 A0A224XJI0 A0A224X6R6 A0A023F0I3 A0A2S2N6Q7 A0A224X724 X1WUG2 A0A2S2PJG2 A0A2S2P698 A0A2S2NH02 A0A224X6U5 A0A224XIB0 A0A224X6W2 A0A224X6S1 A0A2S2P6S3 A0A224XI99 A0A2S2NPJ7 A0A2S2R9D4 A0A2H8TKI9 A0A2S2PHL7 A0A2H8TF37 A0A0J7K3N0 A0A2S2P6C4 A0A023F069 A0A2S2PUS3 A0A2S2QDN9 A0A2S2P7B2 A0A2S2P3X8 A0A1Y1MT28 A0A146KRW8 J9KE07 J9M0H4 A0A2S2NKC3 A0A146KV64 A0A224XK92 A0A1B6C2V8 J9KSK5 A0A2S2QEE0 Q9NBX4 A0A2S2PA10 X1XLN5 A0A2S2NPG3 J9KWJ0 A0A224X7Z3 J9M4H5 A0A2S2N9P4 X1WVQ4 J9LMA3 A0A2S2PBB4 J9KLB1 A0A2S2Q2R8
EC Number
2.7.7.49
Pubmed
EMBL
LBMM01013518
KMQ85605.1
NEVH01007402
PNF35856.1
NEVH01016340
PNF25410.1
+ More
NEVH01015305 PNF27092.1 NEVH01021925 PNF19341.1 NEVH01006736 PNF36716.1 NEVH01008277 PNF34262.1 NEVH01001347 PNF42959.1 NEVH01023979 PNF17579.1 NEVH01020940 PNF20481.1 NEVH01016302 PNF25823.1 NWSH01000032 PCG80496.1 NWSH01007753 PCG62823.1 NEVH01019963 PNF22185.1 LBMM01014395 KMQ85192.1 NEVH01026386 PNF14434.1 ABLF02041861 GEZM01084969 JAV60363.1 GEZM01053022 JAV74219.1 GBHO01021814 GBHO01010549 JAG21790.1 JAG33055.1 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 NEVH01005295 PNF38784.1 LBMM01012078 KMQ86419.1 KU543673 AMS38352.1 GEDC01000337 JAS36961.1 KU543687 AMS38379.1 NEVH01020963 PNF20308.1 GAKP01017773 JAC41179.1 AY508487 AAS13459.1 X17551 CAA35587.1 LBMM01012041 KMQ86458.1 KU543685 AMS38375.1 AY047531 AAK77263.1 CP012527 ALC48130.1 M17214 AAA28508.1 GFTR01008261 JAW08165.1 DNOY01000259 HBE18313.1 GFDF01007661 JAV06423.1 LBMM01006314 KMQ90710.1 GBGD01000389 JAC88500.1 GFTR01008254 JAW08172.1 GECL01003538 JAP02586.1 GGMR01003407 MBY16026.1 GBHO01008295 JAG35309.1 GGMR01000676 MBY13295.1 GFTR01008151 JAW08275.1 NEVH01006721 PNF37225.1 GFTR01008257 JAW08169.1 GFTR01008269 JAW08157.1 GBBI01003747 JAC14965.1 GGMR01000226 MBY12845.1 GFTR01008169 JAW08257.1 ABLF02016613 GGMR01016905 MBY29524.1 GGMR01012305 MBY24924.1 GGMR01003864 MBY16483.1 GFTR01008251 JAW08175.1 GFTR01008246 JAW08180.1 GFTR01008260 JAW08166.1 GFTR01008271 JAW08155.1 GGMR01012443 MBY25062.1 GFTR01008256 JAW08170.1 GGMR01006491 MBY19110.1 GGMS01017365 MBY86568.1 GFXV01002840 MBW14645.1 GGMR01016296 MBY28915.1 GFXV01000908 MBW12713.1 LBMM01015125 KMQ84919.1 GGMR01012283 MBY24902.1 GBBI01003810 JAC14902.1 GGMR01020516 MBY33135.1 GGMS01006655 MBY75858.1 GGMR01012676 MBY25295.1 GGMR01011511 MBY24130.1 GEZM01021881 JAV88842.1 GDHC01020244 JAP98384.1 ABLF02024784 ABLF02024792 ABLF02034460 GGMR01005022 MBY17641.1 GDHC01018248 JAQ00381.1 GFTR01007997 JAW08429.1 GEDC01029594 JAS07704.1 ABLF02014118 ABLF02014121 ABLF02054790 GGMS01006359 MBY75562.1 AF237761 AAF81411.1 GGMR01013409 MBY26028.1 ABLF02017755 ABLF02044739 ABLF02063495 ABLF02065698 ABLF02066197 GGMR01006408 MBY19027.1 ABLF02021819 ABLF02021820 ABLF02055326 GFTR01007929 JAW08497.1 ABLF02017668 ABLF02021394 ABLF02055728 GGMR01000857 MBY13476.1 ABLF02023279 ABLF02041474 ABLF02025048 ABLF02063033 GGMR01014053 MBY26672.1 ABLF02040174 ABLF02040183 ABLF02055266 GGMS01002786 MBY71989.1
NEVH01015305 PNF27092.1 NEVH01021925 PNF19341.1 NEVH01006736 PNF36716.1 NEVH01008277 PNF34262.1 NEVH01001347 PNF42959.1 NEVH01023979 PNF17579.1 NEVH01020940 PNF20481.1 NEVH01016302 PNF25823.1 NWSH01000032 PCG80496.1 NWSH01007753 PCG62823.1 NEVH01019963 PNF22185.1 LBMM01014395 KMQ85192.1 NEVH01026386 PNF14434.1 ABLF02041861 GEZM01084969 JAV60363.1 GEZM01053022 JAV74219.1 GBHO01021814 GBHO01010549 JAG21790.1 JAG33055.1 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 NEVH01005295 PNF38784.1 LBMM01012078 KMQ86419.1 KU543673 AMS38352.1 GEDC01000337 JAS36961.1 KU543687 AMS38379.1 NEVH01020963 PNF20308.1 GAKP01017773 JAC41179.1 AY508487 AAS13459.1 X17551 CAA35587.1 LBMM01012041 KMQ86458.1 KU543685 AMS38375.1 AY047531 AAK77263.1 CP012527 ALC48130.1 M17214 AAA28508.1 GFTR01008261 JAW08165.1 DNOY01000259 HBE18313.1 GFDF01007661 JAV06423.1 LBMM01006314 KMQ90710.1 GBGD01000389 JAC88500.1 GFTR01008254 JAW08172.1 GECL01003538 JAP02586.1 GGMR01003407 MBY16026.1 GBHO01008295 JAG35309.1 GGMR01000676 MBY13295.1 GFTR01008151 JAW08275.1 NEVH01006721 PNF37225.1 GFTR01008257 JAW08169.1 GFTR01008269 JAW08157.1 GBBI01003747 JAC14965.1 GGMR01000226 MBY12845.1 GFTR01008169 JAW08257.1 ABLF02016613 GGMR01016905 MBY29524.1 GGMR01012305 MBY24924.1 GGMR01003864 MBY16483.1 GFTR01008251 JAW08175.1 GFTR01008246 JAW08180.1 GFTR01008260 JAW08166.1 GFTR01008271 JAW08155.1 GGMR01012443 MBY25062.1 GFTR01008256 JAW08170.1 GGMR01006491 MBY19110.1 GGMS01017365 MBY86568.1 GFXV01002840 MBW14645.1 GGMR01016296 MBY28915.1 GFXV01000908 MBW12713.1 LBMM01015125 KMQ84919.1 GGMR01012283 MBY24902.1 GBBI01003810 JAC14902.1 GGMR01020516 MBY33135.1 GGMS01006655 MBY75858.1 GGMR01012676 MBY25295.1 GGMR01011511 MBY24130.1 GEZM01021881 JAV88842.1 GDHC01020244 JAP98384.1 ABLF02024784 ABLF02024792 ABLF02034460 GGMR01005022 MBY17641.1 GDHC01018248 JAQ00381.1 GFTR01007997 JAW08429.1 GEDC01029594 JAS07704.1 ABLF02014118 ABLF02014121 ABLF02054790 GGMS01006359 MBY75562.1 AF237761 AAF81411.1 GGMR01013409 MBY26028.1 ABLF02017755 ABLF02044739 ABLF02063495 ABLF02065698 ABLF02066197 GGMR01006408 MBY19027.1 ABLF02021819 ABLF02021820 ABLF02055326 GFTR01007929 JAW08497.1 ABLF02017668 ABLF02021394 ABLF02055728 GGMR01000857 MBY13476.1 ABLF02023279 ABLF02041474 ABLF02025048 ABLF02063033 GGMR01014053 MBY26672.1 ABLF02040174 ABLF02040183 ABLF02055266 GGMS01002786 MBY71989.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A0J7MYU2
A0A2J7R4U3
A0A2J7Q9Z1
A0A2J7QES2
A0A2J7PSM7
A0A2J7R7D0
+ More
A0A2J7R098 A0A2J7RQ52 A0A2J7PMN4 A0A2J7PVX3 A0A2J7QB68 A0A2A4K918 A0A2A4IUM2 A0A2J7Q0S7 A0A0J7K4E0 A0A2J7PDM0 J9LYS0 A0A1Y1KFR3 A0A1Y1LKN6 A0A0A9YU04 A0A0A9YJ00 A0A2J7RD70 A0A0J7K811 A0A142LX26 A0A1B6EGD8 A0A142LX53 A0A2J7PVE7 A0A034VFD2 Q6R811 Q04135 A0A0J7K7J8 A0A142LX49 Q961V7 A0A0M4EWX7 Q24335 A0A224X6T4 A0A352X8G4 A0A1L8DIZ6 A0A0J7NDW4 A0A069DX78 A0A224XHW8 A0A0V0G5B6 A0A2S2NGV8 A0A0A9YSX4 A0A2S2N814 A0A224X746 A0A2J7R8R8 A0A224XJI0 A0A224X6R6 A0A023F0I3 A0A2S2N6Q7 A0A224X724 X1WUG2 A0A2S2PJG2 A0A2S2P698 A0A2S2NH02 A0A224X6U5 A0A224XIB0 A0A224X6W2 A0A224X6S1 A0A2S2P6S3 A0A224XI99 A0A2S2NPJ7 A0A2S2R9D4 A0A2H8TKI9 A0A2S2PHL7 A0A2H8TF37 A0A0J7K3N0 A0A2S2P6C4 A0A023F069 A0A2S2PUS3 A0A2S2QDN9 A0A2S2P7B2 A0A2S2P3X8 A0A1Y1MT28 A0A146KRW8 J9KE07 J9M0H4 A0A2S2NKC3 A0A146KV64 A0A224XK92 A0A1B6C2V8 J9KSK5 A0A2S2QEE0 Q9NBX4 A0A2S2PA10 X1XLN5 A0A2S2NPG3 J9KWJ0 A0A224X7Z3 J9M4H5 A0A2S2N9P4 X1WVQ4 J9LMA3 A0A2S2PBB4 J9KLB1 A0A2S2Q2R8
A0A2J7R098 A0A2J7RQ52 A0A2J7PMN4 A0A2J7PVX3 A0A2J7QB68 A0A2A4K918 A0A2A4IUM2 A0A2J7Q0S7 A0A0J7K4E0 A0A2J7PDM0 J9LYS0 A0A1Y1KFR3 A0A1Y1LKN6 A0A0A9YU04 A0A0A9YJ00 A0A2J7RD70 A0A0J7K811 A0A142LX26 A0A1B6EGD8 A0A142LX53 A0A2J7PVE7 A0A034VFD2 Q6R811 Q04135 A0A0J7K7J8 A0A142LX49 Q961V7 A0A0M4EWX7 Q24335 A0A224X6T4 A0A352X8G4 A0A1L8DIZ6 A0A0J7NDW4 A0A069DX78 A0A224XHW8 A0A0V0G5B6 A0A2S2NGV8 A0A0A9YSX4 A0A2S2N814 A0A224X746 A0A2J7R8R8 A0A224XJI0 A0A224X6R6 A0A023F0I3 A0A2S2N6Q7 A0A224X724 X1WUG2 A0A2S2PJG2 A0A2S2P698 A0A2S2NH02 A0A224X6U5 A0A224XIB0 A0A224X6W2 A0A224X6S1 A0A2S2P6S3 A0A224XI99 A0A2S2NPJ7 A0A2S2R9D4 A0A2H8TKI9 A0A2S2PHL7 A0A2H8TF37 A0A0J7K3N0 A0A2S2P6C4 A0A023F069 A0A2S2PUS3 A0A2S2QDN9 A0A2S2P7B2 A0A2S2P3X8 A0A1Y1MT28 A0A146KRW8 J9KE07 J9M0H4 A0A2S2NKC3 A0A146KV64 A0A224XK92 A0A1B6C2V8 J9KSK5 A0A2S2QEE0 Q9NBX4 A0A2S2PA10 X1XLN5 A0A2S2NPG3 J9KWJ0 A0A224X7Z3 J9M4H5 A0A2S2N9P4 X1WVQ4 J9LMA3 A0A2S2PBB4 J9KLB1 A0A2S2Q2R8
PDB
1WDU
E-value=0.00225748,
Score=100
Ontologies
KEGG
Topology
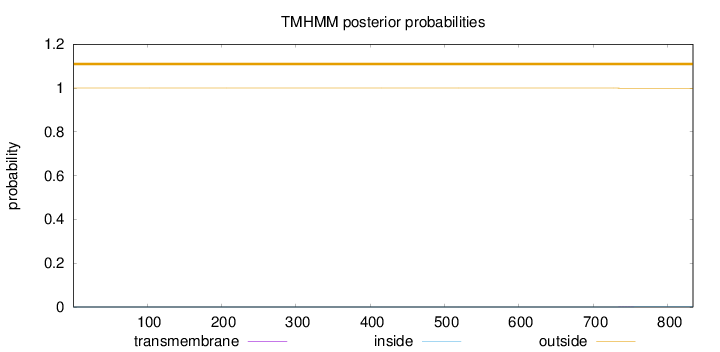
Length:
834
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01699
Exp number, first 60 AAs:
0.00072
Total prob of N-in:
0.00005
outside
1 - 834
Population Genetic Test Statistics
Pi
60.160686
Theta
150.554297
Tajima's D
-1.186091
CLR
140.585461
CSRT
0.104994750262487
Interpretation
Uncertain
