Gene
KWMTBOMO00928
Pre Gene Modal
BGIBMGA009645
Annotation
PREDICTED:_zinc_transporter_foi-like_[Bombyx_mori]
Full name
Zinc transporter foi
Alternative Name
Protein fear-of-intimacy
Protein kastchen
Protein kastchen
Location in the cell
PlasmaMembrane Reliability : 1.768
Sequence
CDS
ATGTCACATCATTTGGTGACAGTTTGCATGTTTTGTCTACTGTGCGCCGCCCACACTTGCGGCTCCCACGTGGACATACAGACAGATGGAGTCTCGGCACACGAACCGAGGCTCGAGACCACGGACCACATGCCCCTCGAATTAGAAGAGAAAAGGAGGCTGGATTCGAGGAACCGCTACCATCAGAAGCTTCAGAAGATCAGACAGAAGGATCGCTTGAAGAGGGACGCGAAGAATGTCGACCCTGAGTTGTATGTAGCCCAGATATTCGGCATGTACGGGGACGCCGCTAGTATGACCATGAACCTGACCGGCTTCAACAAGATGCTTGAAGAGCTAGACCTGCATAAGCTGATCGATGGTGGCCAAACGACCGACAGCACGGCCTACTTCCACGTTCAGACGTCGAAGGATGAAGAAGTTGTCAGCTGCGTTAACAGTTCAGAACTGGTTACAACGGTGAACACGAGGTTAAAGCCCCACGACCATGGCCACGGTCACGAGGATCACACCCACAGCGACCACGACCACGAACTCCTAATATCCAAAGACACGTTGAGGTCAGTCTGTCCAATATTGCTGTACCAGAAACTAGCAAACACGTCGATAGAACGTTCTGGATGCGTCAAAGAAACGAATCTACTGGTTCGCCACCACCGAGACTTGGAAGAGAAGGAGAACGCGTACACGAAAAACATGTTCGCGGTCTGGCTGTATTCATCCCTGAGCATAACCGCGATCAGCGCCTGTGGTCTCCTAGGCGTGGCGGTCATCCCTGTCATGCACAAGACTTATTATAACCATCTGCTGCAGTTTCTGGTGGCTCTGGCCGTAGGGACGCTGTGCGGGGACGCCTTGCTGCACCTGATGCCTCACGCGATGAGTCCCATGCACGACCACGAGGGCGGTGGACATGCGGTGGCGGAAATGTCGCACGACGACGGAATGTGGAAGGGATTCGCCGCCATGCTGGGCGTTGTGTTCTTCTATTTCACAGAGAAGGGATTAACGGTTGTCGTTGAGTGGAGGAAACGCAGGCAGAAGTTGGAGGAAGACAAACTTCCGACCAGAGTGCGCGTCCTTAAGGACGAGACGGTCGGGGGATGTTCGGGTGGTTCAAAAGCTCCCATAACGAACACAACTTCGAACAGTCTCATGAAGATCTTTAAGCGGGACGAAAAGGGAGATCCCACAACGAAAACTTGCAAGCACAAATACTCTGAATATCCTTATTGTTATGATGAAATCGATACGGATACTCACGACGAGCACCATCTGAGGGACGGTATACCGCCCAGCCCTAAAGCGAAGCACAACTCCGTAAGCGTGGTCTCGTCCAACGCCCTCAATCCTGAGGGCAAAGAGGAACCCACAGCCAAGGACTGGCTGCTGAAAGCCGAGACCACTCCTGCTGTTGTCGAAATGAACAATATCAAACACAAAGAAGACGGGTCCAAGGATCTGAAGGATGGCGGCAGTTACACAGTTATATTGAGGGAACATTCTAGAGCGCACCACGGTCACTCCCATGCCCATGGGCACGTGCACGCTCCACCGTCGTCGATAGCGTCCGTGGCCTGGATGGTGATCATGGGTGACGGCCTCCACAATTTCACAGACGGCATGGCCATTGGCGCAGCGTTCGCATCCAGCATAGCAGGTGGATTCTCGACCGCCATCGCGGTGCTCTGCCATGAACTTCCTCACGAGCTGGGCGATTTCGCGGTGTTACTGAAGGCGGGCATGTCTGTTAAGCGTGCTGTCTGCTACAACATACTGTCGTCGGCTCTGTGCTTGGCGGGGATGGTCTGCGGGGTCCTGGCCGGGCACGCCCCCTCGGCAACCAGGTGGCTATTCGCAGTGGCAGCTGGCATGTTCCTCTACATAGCCCTCGTTGATATGATGCCTGAACTTAGCACGTCTCACAGCAACGAGGGTACGCTATGCCAATGTGTGCTGCAACTGATGGGTCTAGCCAGCGGCATCGGTATAATGCTGGTCATCGCTCTATACGAAGACGACCTCAAAAACCTGTTCGGCTGA
Protein
MSHHLVTVCMFCLLCAAHTCGSHVDIQTDGVSAHEPRLETTDHMPLELEEKRRLDSRNRYHQKLQKIRQKDRLKRDAKNVDPELYVAQIFGMYGDAASMTMNLTGFNKMLEELDLHKLIDGGQTTDSTAYFHVQTSKDEEVVSCVNSSELVTTVNTRLKPHDHGHGHEDHTHSDHDHELLISKDTLRSVCPILLYQKLANTSIERSGCVKETNLLVRHHRDLEEKENAYTKNMFAVWLYSSLSITAISACGLLGVAVIPVMHKTYYNHLLQFLVALAVGTLCGDALLHLMPHAMSPMHDHEGGGHAVAEMSHDDGMWKGFAAMLGVVFFYFTEKGLTVVVEWRKRRQKLEEDKLPTRVRVLKDETVGGCSGGSKAPITNTTSNSLMKIFKRDEKGDPTTKTCKHKYSEYPYCYDEIDTDTHDEHHLRDGIPPSPKAKHNSVSVVSSNALNPEGKEEPTAKDWLLKAETTPAVVEMNNIKHKEDGSKDLKDGGSYTVILREHSRAHHGHSHAHGHVHAPPSSIASVAWMVIMGDGLHNFTDGMAIGAAFASSIAGGFSTAIAVLCHELPHELGDFAVLLKAGMSVKRAVCYNILSSALCLAGMVCGVLAGHAPSATRWLFAVAAGMFLYIALVDMMPELSTSHSNEGTLCQCVLQLMGLASGIGIMLVIALYEDDLKNLFG
Summary
Description
Required for the normal migration of longitudinal and peripheral glial cells. During larval development, required for the migration of the subretinal glia into the eye disk. During embryonic development, also controls the migration of muscle cells toward their attachment sites. Required in the mesoderm for the correct morphogenesis of embryonic gonad and for tracheal branch fusion during tracheal development. Shg may be cooperating with foi to mediate a common mechanism for gonad and tracheal morphogenesis. Acts as a zinc transporter in both yeast and mammalian cells.
Similarity
Belongs to the ZIP transporter (TC 2.A.5) family.
Keywords
Cell membrane
Complete proteome
Developmental protein
Differentiation
Glycoprotein
Ion transport
Membrane
Neurogenesis
Phosphoprotein
Reference proteome
Signal
Transmembrane
Transmembrane helix
Transport
Zinc
Zinc transport
Feature
chain Zinc transporter foi
Uniprot
A0A2H1V9Z0
A0A212FBM1
A0A0N0PD99
A0A194PT92
A0A2A4J0W5
A0A0L7LFI5
+ More
A0A1B0CGU2 A0A1L8DDX3 A0A1L8DE56 A0A1L8DDW2 A0A1L8DDY9 E2B260 A0A0C9R9D7 A0A195C9I3 A0A151I5J1 A0A1B6GI83 A0A1B6KB35 A0A0M8ZNQ0 A0A2A3EMP4 A0A088AC68 A0A151WNG3 A0A232EWT8 A0A158NU01 A0A0L7RET1 F4WXB0 A0A026X0W0 A0A336LX90 A0A195DJG1 A0A1B6F1U2 A0A3L8DST0 A0A1B6DE89 A0A1J1JA36 A0A182M706 A0A1W4XTK0 A0A154P2S3 A0A2J7QRD8 A0A2J7QRD0 A0A1Y1KCT6 A0A2J7QRE2 A0A2J7QRF0 A0A224XI10 A0A1Q3FVT7 A0A1Q3FVM2 A0A310STU8 A0A0P4VRX1 Q16RN0 A0A182QQ70 A0A139WEH4 A0A182WCK3 A0A182RDF0 A0A084VDG5 A0A182S8G6 A0A182K0Z9 A0A182PJW2 B4PG09 A0A0A9XM25 B4N5A7 Q29R33 M9PES6 Q9VSL7
A0A1B0CGU2 A0A1L8DDX3 A0A1L8DE56 A0A1L8DDW2 A0A1L8DDY9 E2B260 A0A0C9R9D7 A0A195C9I3 A0A151I5J1 A0A1B6GI83 A0A1B6KB35 A0A0M8ZNQ0 A0A2A3EMP4 A0A088AC68 A0A151WNG3 A0A232EWT8 A0A158NU01 A0A0L7RET1 F4WXB0 A0A026X0W0 A0A336LX90 A0A195DJG1 A0A1B6F1U2 A0A3L8DST0 A0A1B6DE89 A0A1J1JA36 A0A182M706 A0A1W4XTK0 A0A154P2S3 A0A2J7QRD8 A0A2J7QRD0 A0A1Y1KCT6 A0A2J7QRE2 A0A2J7QRF0 A0A224XI10 A0A1Q3FVT7 A0A1Q3FVM2 A0A310STU8 A0A0P4VRX1 Q16RN0 A0A182QQ70 A0A139WEH4 A0A182WCK3 A0A182RDF0 A0A084VDG5 A0A182S8G6 A0A182K0Z9 A0A182PJW2 B4PG09 A0A0A9XM25 B4N5A7 Q29R33 M9PES6 Q9VSL7
Pubmed
22118469
26354079
26227816
20798317
28648823
21347285
+ More
21719571 24508170 30249741 28004739 27129103 17510324 18362917 19820115 24438588 17994087 17550304 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12702650 15464587 15509557 18327897
21719571 24508170 30249741 28004739 27129103 17510324 18362917 19820115 24438588 17994087 17550304 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12702650 15464587 15509557 18327897
EMBL
ODYU01001445
SOQ37655.1
AGBW02009306
OWR51113.1
KQ460280
KPJ16311.1
+ More
KQ459593 KPI96636.1 NWSH01003798 PCG65807.1 JTDY01001278 KOB74313.1 AJWK01011588 AJWK01011589 AJWK01011590 AJWK01011591 GFDF01009428 JAV04656.1 GFDF01009427 JAV04657.1 GFDF01009425 JAV04659.1 GFDF01009426 JAV04658.1 GL445059 EFN60235.1 GBYB01013018 GBYB01013019 JAG82785.1 JAG82786.1 KQ978143 KYM96773.1 KQ976445 KYM86078.1 GECZ01007617 JAS62152.1 GEBQ01031318 GEBQ01029085 GEBQ01028066 GEBQ01007362 JAT08659.1 JAT10892.1 JAT11911.1 JAT32615.1 KQ435970 KOX67797.1 KZ288210 PBC32960.1 KQ982926 KYQ49225.1 NNAY01001826 OXU22795.1 ADTU01026230 KQ414608 KOC69467.1 GL888423 EGI61159.1 KK107046 EZA61713.1 UFQT01000031 SSX18258.1 KQ980794 KYN13035.1 GECZ01025631 JAS44138.1 QOIP01000004 RLU23386.1 GEDC01013280 JAS24018.1 CVRI01000075 CRL08732.1 AXCM01001750 KQ434796 KZC05644.1 NEVH01011907 PNF31148.1 PNF31149.1 GEZM01086504 JAV59279.1 PNF31146.1 PNF31147.1 GFTR01006978 JAW09448.1 GFDL01003387 JAV31658.1 GFDL01003391 JAV31654.1 KQ760512 OAD60051.1 GDKW01002775 JAI53820.1 CH477702 EAT37058.1 AXCN02000347 KQ971354 KYB26289.1 ATLV01011335 KE524664 KFB36009.1 CM000159 EDW93153.1 KRK00986.1 GBHO01023731 GBHO01023730 GBRD01014230 GDHC01016407 JAG19873.1 JAG19874.1 JAG51596.1 JAQ02222.1 CH964101 EDW79546.1 BT024207 ABC86269.1 AE014296 AGB94279.1 AGB94280.1 AHN58010.1 AAF50401.3
KQ459593 KPI96636.1 NWSH01003798 PCG65807.1 JTDY01001278 KOB74313.1 AJWK01011588 AJWK01011589 AJWK01011590 AJWK01011591 GFDF01009428 JAV04656.1 GFDF01009427 JAV04657.1 GFDF01009425 JAV04659.1 GFDF01009426 JAV04658.1 GL445059 EFN60235.1 GBYB01013018 GBYB01013019 JAG82785.1 JAG82786.1 KQ978143 KYM96773.1 KQ976445 KYM86078.1 GECZ01007617 JAS62152.1 GEBQ01031318 GEBQ01029085 GEBQ01028066 GEBQ01007362 JAT08659.1 JAT10892.1 JAT11911.1 JAT32615.1 KQ435970 KOX67797.1 KZ288210 PBC32960.1 KQ982926 KYQ49225.1 NNAY01001826 OXU22795.1 ADTU01026230 KQ414608 KOC69467.1 GL888423 EGI61159.1 KK107046 EZA61713.1 UFQT01000031 SSX18258.1 KQ980794 KYN13035.1 GECZ01025631 JAS44138.1 QOIP01000004 RLU23386.1 GEDC01013280 JAS24018.1 CVRI01000075 CRL08732.1 AXCM01001750 KQ434796 KZC05644.1 NEVH01011907 PNF31148.1 PNF31149.1 GEZM01086504 JAV59279.1 PNF31146.1 PNF31147.1 GFTR01006978 JAW09448.1 GFDL01003387 JAV31658.1 GFDL01003391 JAV31654.1 KQ760512 OAD60051.1 GDKW01002775 JAI53820.1 CH477702 EAT37058.1 AXCN02000347 KQ971354 KYB26289.1 ATLV01011335 KE524664 KFB36009.1 CM000159 EDW93153.1 KRK00986.1 GBHO01023731 GBHO01023730 GBRD01014230 GDHC01016407 JAG19873.1 JAG19874.1 JAG51596.1 JAQ02222.1 CH964101 EDW79546.1 BT024207 ABC86269.1 AE014296 AGB94279.1 AGB94280.1 AHN58010.1 AAF50401.3
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000037510
UP000092461
+ More
UP000000311 UP000078542 UP000078540 UP000053105 UP000242457 UP000005203 UP000075809 UP000215335 UP000005205 UP000053825 UP000007755 UP000053097 UP000078492 UP000279307 UP000183832 UP000075883 UP000192223 UP000076502 UP000235965 UP000008820 UP000075886 UP000007266 UP000075920 UP000075900 UP000030765 UP000075901 UP000075881 UP000075885 UP000002282 UP000007798 UP000000803
UP000000311 UP000078542 UP000078540 UP000053105 UP000242457 UP000005203 UP000075809 UP000215335 UP000005205 UP000053825 UP000007755 UP000053097 UP000078492 UP000279307 UP000183832 UP000075883 UP000192223 UP000076502 UP000235965 UP000008820 UP000075886 UP000007266 UP000075920 UP000075900 UP000030765 UP000075901 UP000075881 UP000075885 UP000002282 UP000007798 UP000000803
SUPFAM
SSF49899
SSF49899
ProteinModelPortal
A0A2H1V9Z0
A0A212FBM1
A0A0N0PD99
A0A194PT92
A0A2A4J0W5
A0A0L7LFI5
+ More
A0A1B0CGU2 A0A1L8DDX3 A0A1L8DE56 A0A1L8DDW2 A0A1L8DDY9 E2B260 A0A0C9R9D7 A0A195C9I3 A0A151I5J1 A0A1B6GI83 A0A1B6KB35 A0A0M8ZNQ0 A0A2A3EMP4 A0A088AC68 A0A151WNG3 A0A232EWT8 A0A158NU01 A0A0L7RET1 F4WXB0 A0A026X0W0 A0A336LX90 A0A195DJG1 A0A1B6F1U2 A0A3L8DST0 A0A1B6DE89 A0A1J1JA36 A0A182M706 A0A1W4XTK0 A0A154P2S3 A0A2J7QRD8 A0A2J7QRD0 A0A1Y1KCT6 A0A2J7QRE2 A0A2J7QRF0 A0A224XI10 A0A1Q3FVT7 A0A1Q3FVM2 A0A310STU8 A0A0P4VRX1 Q16RN0 A0A182QQ70 A0A139WEH4 A0A182WCK3 A0A182RDF0 A0A084VDG5 A0A182S8G6 A0A182K0Z9 A0A182PJW2 B4PG09 A0A0A9XM25 B4N5A7 Q29R33 M9PES6 Q9VSL7
A0A1B0CGU2 A0A1L8DDX3 A0A1L8DE56 A0A1L8DDW2 A0A1L8DDY9 E2B260 A0A0C9R9D7 A0A195C9I3 A0A151I5J1 A0A1B6GI83 A0A1B6KB35 A0A0M8ZNQ0 A0A2A3EMP4 A0A088AC68 A0A151WNG3 A0A232EWT8 A0A158NU01 A0A0L7RET1 F4WXB0 A0A026X0W0 A0A336LX90 A0A195DJG1 A0A1B6F1U2 A0A3L8DST0 A0A1B6DE89 A0A1J1JA36 A0A182M706 A0A1W4XTK0 A0A154P2S3 A0A2J7QRD8 A0A2J7QRD0 A0A1Y1KCT6 A0A2J7QRE2 A0A2J7QRF0 A0A224XI10 A0A1Q3FVT7 A0A1Q3FVM2 A0A310STU8 A0A0P4VRX1 Q16RN0 A0A182QQ70 A0A139WEH4 A0A182WCK3 A0A182RDF0 A0A084VDG5 A0A182S8G6 A0A182K0Z9 A0A182PJW2 B4PG09 A0A0A9XM25 B4N5A7 Q29R33 M9PES6 Q9VSL7
Ontologies
GO
GO:0046873
GO:0016021
GO:0005509
GO:0006882
GO:0005385
GO:0071578
GO:0005887
GO:0005537
GO:0005737
GO:0006888
GO:0007506
GO:0035262
GO:0009986
GO:0007417
GO:0008354
GO:0016323
GO:0035147
GO:0030154
GO:0005886
GO:0006829
GO:0007280
GO:0016477
GO:0008406
GO:0016020
GO:0030001
GO:0006422
GO:0004812
GO:0006418
GO:0004887
GO:0003707
GO:0055085
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 21,
Likelihood: 0.987042
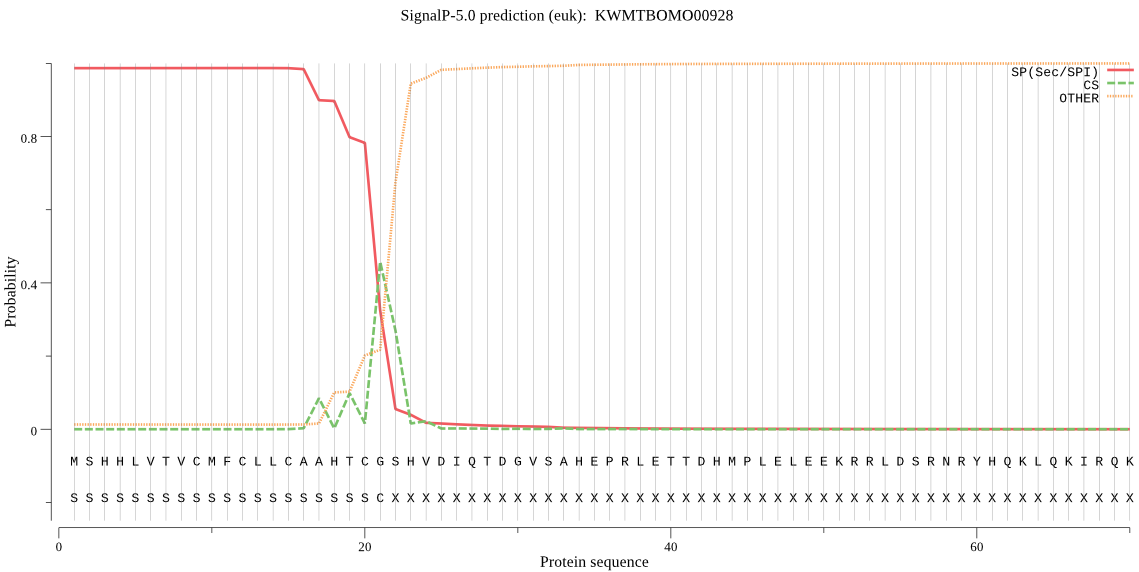
Length:
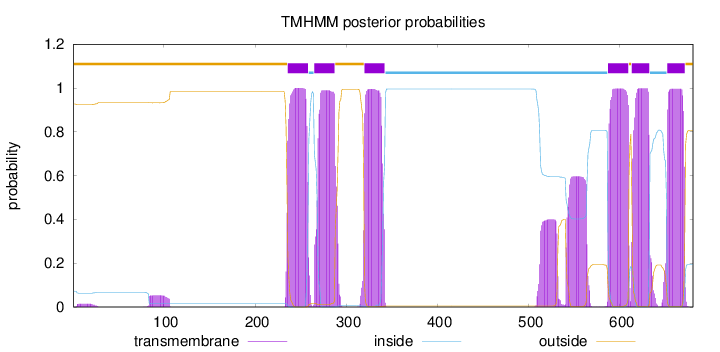
680
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
151.69237
Exp number, first 60 AAs:
0.29013
Total prob of N-in:
0.07273
outside
1 - 235
TMhelix
236 - 258
inside
259 - 264
TMhelix
265 - 287
outside
288 - 319
TMhelix
320 - 342
inside
343 - 586
TMhelix
587 - 609
outside
610 - 612
TMhelix
613 - 632
inside
633 - 651
TMhelix
652 - 671
outside
672 - 680
Population Genetic Test Statistics
Pi
209.145984
Theta
203.05214
Tajima's D
0.216032
CLR
0.526011
CSRT
0.430678466076696
Interpretation
Uncertain
