Gene
KWMTBOMO00922
Pre Gene Modal
BGIBMGA009791
Annotation
carotenoid-binding_protein_isoform_1_[Bombyx_mori]
Full name
Steroidogenic acute regulatory protein-like
Location in the cell
PlasmaMembrane Reliability : 3.266
Sequence
CDS
ATGTCTCATCCTCACATACGAGAAGCAGCTGAATCTCTCTTAAACAGCGTCACGTCCCCGCCTTATAGAATGATGAATCATTCACAGTCCGTGAATCTGATGACCGAGGACCTAGTGGCGGGACACAGGCCCGCCGGGAAGATGTCCTCGGTCAGAAGGTTCTTCTGTCTCTTCGTGACCTTCGATTTAATGTTCACAAGCCTCATGTGGCTGATTTGTGTTATGGTTCGTAACGAGACGTTAGACCAAATATTGCAAAGAGAAATTATTAGTTATAATATAAAACTATCGTTATTCGACATAGTGGTGTTAGCTGTGATAAGATTTACAATATTATTGTTGTTCTACGCTTTATTATATATCAACAATTGGAGTGTGATAGCTCTCTCGACAAGCGGCACTTGCGCCTTTTTAATAGCGAAAGTGTTCGTATTCGTTTGGTCCGATGCGTCACAGCCCGTGTATCAAGTGTTCCTTATACTGACTTCGTTTACCTTGTCATGGGGAGAGGCCTGGTTCTTGGACTTCAGGGTCTTACCGCAGGAGCTACACTGCAAGGAGATACTAGATTCCTTAGCTCAGCACAGCGTCTCCGAGCGCACGCCGCTACTGGACCCTCAGCCCGGTGTGAATTCGACGTACGCAGAGTCCACGGTTAAATGGTTCTCCCCGGTCGACACGCCGGAGTCGAGTCCACGGCCGCGCAGGTTCGGGGAGCAAGTCATATTGACGCAGGAGCAGATCGATGAGTACAAGAGTCAGGCTAACGAGAGCATGGCGAACGCTTGGCGCATCATCACGCTGCCAAACTGGACGGTGGAGAAGCGAGGGTCCGTCAGGGGAGACGTCGTGGAGTCCAGGAAAGTTGAAGGCTACGGCAAGGTCTATAGATTTACGGGCGTCGTCAACTGTCCCGCCAGGTTTCTATACGAGGAGTTCAAGAACAACCTGACGAAGTTGCCCGAATGGAATCCGACGATATTGAAATGCGAAATAATTAAGGAAATCGGCGATGGCGTGGACCTGTCGTACCAGGTGACGGCTGGCGGGGGACGAGGCATAATAACCCCTCGCGACTTCGTGATCCTGCGCCGGATCGCGCTACTCTCCCGGGAGGGACGCGTCGTCGACGACAATCCCCACGGATACATCAGTAGCGGAGTCAGCGTTCAAGTTCCCGGGTACCCGCCCCTCAAGGAGATGGTTAGGGGACACAATAAAGTTGGTTGCTGGTACCTACAGCCTCGGACTGTACAAACTGCTGGAGGAAAAATCGAGGACCAGACCCTGTTCCAATGGTTGATGTGTTGTGATCTCAAAGGTAAAATACCTCAATTCGTGTTGGACGTCGCTTTCGCCACTGTGATGCTGGACTACATCGTGCACGTCAGGAAGTTCGTGGCCGAAGCGAAGGCCAGAGCCGAATTCTGA
Protein
MSHPHIREAAESLLNSVTSPPYRMMNHSQSVNLMTEDLVAGHRPAGKMSSVRRFFCLFVTFDLMFTSLMWLICVMVRNETLDQILQREIISYNIKLSLFDIVVLAVIRFTILLLFYALLYINNWSVIALSTSGTCAFLIAKVFVFVWSDASQPVYQVFLILTSFTLSWGEAWFLDFRVLPQELHCKEILDSLAQHSVSERTPLLDPQPGVNSTYAESTVKWFSPVDTPESSPRPRRFGEQVILTQEQIDEYKSQANESMANAWRIITLPNWTVEKRGSVRGDVVESRKVEGYGKVYRFTGVVNCPARFLYEEFKNNLTKLPEWNPTILKCEIIKEIGDGVDLSYQVTAGGGRGIITPRDFVILRRIALLSREGRVVDDNPHGYISSGVSVQVPGYPPLKEMVRGHNKVGCWYLQPRTVQTAGGKIEDQTLFQWLMCCDLKGKIPQFVLDVAFATVMLDYIVHVRKFVAEAKARAEF
Summary
Description
May bind to and transport cholesterol and may play a role in ecdysteroid biosynthesis.
Keywords
Alternative splicing
Complete proteome
Lipid transport
Lipid-binding
Membrane
Reference proteome
Steroidogenesis
Transmembrane
Transmembrane helix
Transport
Feature
chain Steroidogenic acute regulatory protein-like
splice variant In isoform B.
splice variant In isoform B.
Uniprot
Q3V5Y6
D6WT76
A0A2J7RRK7
A0A067RND7
A0A2J7RRL2
A0A2J7RRI0
+ More
A0A2J7RRJ7 E9FUI1 A0A1Y1N158 A0A1Y1MZS7 A0A0P5V2X9 A0A0P5JA07 A0A0P5LQT0 A0A0P6C0B4 A0A0P5B236 A0A0P5CZM4 A0A0N7ZPD8 A0A0P4YZF7 A0A0P5V270 A0A0P4ZIB0 A0A0P5BKX0 V5GWI2 K7INF7 E2BKV3 A0A154PI58 A0A0N8EK28 E2AXF6 A0A232ENV8 A0A0N8CZL7 A0A0C9RE63 A0A189XGA0 A0A3L8DCB3 A0A026WKG3 A0A0L0CLS7 A0A1L8DY45 A0A1L8DYJ2 B4H561 B5E1A0 A0A3B0J3D2 A0A0K8W3V8 A0A0C9R542 A0A0K8UUC4 A0A034VJP0 A0A0M4EB96 A0A3B0JM71 A0A1A9VWE5 A0A195EET5 A0A195D402 A0A1I8PDW8 A0A2P8XXI2 A0A0L7RBS2 A0A195ESU6 A0A158NTM5 B4MP76 A0A151I2E5 A0A182Q372 A0A087ZMS2 A0A1W4W0I6 A0A182WWT2 A0A182HWG1 A0A1A9WA31 A0A151X343 A0A0P4WBR8 T1PBR4 A0A1I8NFT8 A0A0A1XIE6 B3NQU6 A0A1B0B6G6 B4PBD0 A0A182NJQ0 W8BVC6 A0A182Y264 A0A1Q3G1B7 A0A1B0D5P1 B4QC20 A0A1B0FN74 Q9W145-2 A0A1A9YDA3 B4IH40 Q9W145 B4LJI9 A0A1Q3G164
A0A2J7RRJ7 E9FUI1 A0A1Y1N158 A0A1Y1MZS7 A0A0P5V2X9 A0A0P5JA07 A0A0P5LQT0 A0A0P6C0B4 A0A0P5B236 A0A0P5CZM4 A0A0N7ZPD8 A0A0P4YZF7 A0A0P5V270 A0A0P4ZIB0 A0A0P5BKX0 V5GWI2 K7INF7 E2BKV3 A0A154PI58 A0A0N8EK28 E2AXF6 A0A232ENV8 A0A0N8CZL7 A0A0C9RE63 A0A189XGA0 A0A3L8DCB3 A0A026WKG3 A0A0L0CLS7 A0A1L8DY45 A0A1L8DYJ2 B4H561 B5E1A0 A0A3B0J3D2 A0A0K8W3V8 A0A0C9R542 A0A0K8UUC4 A0A034VJP0 A0A0M4EB96 A0A3B0JM71 A0A1A9VWE5 A0A195EET5 A0A195D402 A0A1I8PDW8 A0A2P8XXI2 A0A0L7RBS2 A0A195ESU6 A0A158NTM5 B4MP76 A0A151I2E5 A0A182Q372 A0A087ZMS2 A0A1W4W0I6 A0A182WWT2 A0A182HWG1 A0A1A9WA31 A0A151X343 A0A0P4WBR8 T1PBR4 A0A1I8NFT8 A0A0A1XIE6 B3NQU6 A0A1B0B6G6 B4PBD0 A0A182NJQ0 W8BVC6 A0A182Y264 A0A1Q3G1B7 A0A1B0D5P1 B4QC20 A0A1B0FN74 Q9W145-2 A0A1A9YDA3 B4IH40 Q9W145 B4LJI9 A0A1Q3G164
Pubmed
EMBL
AB211534
AB211535
BAE44461.1
BAE44462.1
KQ971354
EFA07554.1
+ More
NEVH01000603 PNF43452.1 KK852521 KDR22110.1 PNF43454.1 PNF43449.1 PNF43453.1 GL732525 EFX88733.1 GEZM01016737 JAV91158.1 GEZM01016736 JAV91159.1 GDIP01105553 JAL98161.1 GDIQ01228002 GDIQ01228001 JAK23723.1 GDIQ01242450 GDIQ01211903 GDIQ01192960 GDIQ01190910 GDIQ01189508 GDIP01134150 GDIQ01072252 LRGB01000868 JAK60815.1 JAL69564.1 KZS15573.1 GDIP01011478 JAM92237.1 GDIP01190930 JAJ32472.1 GDIP01167391 JAJ56011.1 GDIP01225633 JAI97768.1 GDIP01222784 JAJ00618.1 GDIP01108097 JAL95617.1 GDIP01225632 JAI97769.1 GDIP01188338 JAJ35064.1 GALX01002409 JAB66057.1 GL448826 EFN83702.1 KQ434899 KZC10880.1 GDIQ01019044 JAN75693.1 GL443549 EFN61880.1 NNAY01003047 OXU20059.1 GDIP01083311 JAM20404.1 GBYB01011437 GBYB01011438 JAG81204.1 JAG81205.1 KT072711 ALO17561.1 QOIP01000010 RLU17971.1 KK107171 EZA56111.1 JRES01000204 KNC33323.1 GFDF01002748 JAV11336.1 GFDF01002749 JAV11335.1 CH479210 EDW32897.1 CM000071 EDY69488.1 ENO01925.2 OUUW01000001 SPP73742.1 GDHF01027525 GDHF01010397 GDHF01009665 GDHF01008147 GDHF01006426 GDHF01005581 JAI24789.1 JAI41917.1 JAI42649.1 JAI44167.1 JAI45888.1 JAI46733.1 GBYB01011439 JAG81206.1 GDHF01022186 GDHF01017644 JAI30128.1 JAI34670.1 GAKP01016923 GAKP01016922 GAKP01016921 GAKP01016920 JAC42030.1 CP012524 ALC40752.1 SPP73741.1 KQ978983 KYN26748.1 KQ976881 KYN07617.1 PYGN01001214 PSN36624.1 KQ414617 KOC68377.1 KQ981993 KYN30987.1 ADTU01026048 ADTU01026049 CH963848 EDW73915.2 KQ976543 KYM81019.1 AXCN02000497 APCN01001489 KQ982562 KYQ54846.1 GDRN01054278 JAI66101.1 KA645403 AFP60032.1 GBXI01008943 GBXI01003541 JAD05349.1 JAD10751.1 CH954179 EDV57029.1 JXJN01009092 CM000158 EDW92562.1 KRK00579.1 GAMC01013491 JAB93064.1 GFDL01001453 JAV33592.1 AJVK01025461 AJVK01025462 CM000362 CM002911 EDX08575.1 KMY96376.1 CCAG010014920 AY455866 AE013599 AY102683 AAM27512.1 AAR19767.1 CH480838 EDW49216.1 CH940648 EDW61557.1 KRF80062.1 KRF80063.1 GFDL01001513 JAV33532.1
NEVH01000603 PNF43452.1 KK852521 KDR22110.1 PNF43454.1 PNF43449.1 PNF43453.1 GL732525 EFX88733.1 GEZM01016737 JAV91158.1 GEZM01016736 JAV91159.1 GDIP01105553 JAL98161.1 GDIQ01228002 GDIQ01228001 JAK23723.1 GDIQ01242450 GDIQ01211903 GDIQ01192960 GDIQ01190910 GDIQ01189508 GDIP01134150 GDIQ01072252 LRGB01000868 JAK60815.1 JAL69564.1 KZS15573.1 GDIP01011478 JAM92237.1 GDIP01190930 JAJ32472.1 GDIP01167391 JAJ56011.1 GDIP01225633 JAI97768.1 GDIP01222784 JAJ00618.1 GDIP01108097 JAL95617.1 GDIP01225632 JAI97769.1 GDIP01188338 JAJ35064.1 GALX01002409 JAB66057.1 GL448826 EFN83702.1 KQ434899 KZC10880.1 GDIQ01019044 JAN75693.1 GL443549 EFN61880.1 NNAY01003047 OXU20059.1 GDIP01083311 JAM20404.1 GBYB01011437 GBYB01011438 JAG81204.1 JAG81205.1 KT072711 ALO17561.1 QOIP01000010 RLU17971.1 KK107171 EZA56111.1 JRES01000204 KNC33323.1 GFDF01002748 JAV11336.1 GFDF01002749 JAV11335.1 CH479210 EDW32897.1 CM000071 EDY69488.1 ENO01925.2 OUUW01000001 SPP73742.1 GDHF01027525 GDHF01010397 GDHF01009665 GDHF01008147 GDHF01006426 GDHF01005581 JAI24789.1 JAI41917.1 JAI42649.1 JAI44167.1 JAI45888.1 JAI46733.1 GBYB01011439 JAG81206.1 GDHF01022186 GDHF01017644 JAI30128.1 JAI34670.1 GAKP01016923 GAKP01016922 GAKP01016921 GAKP01016920 JAC42030.1 CP012524 ALC40752.1 SPP73741.1 KQ978983 KYN26748.1 KQ976881 KYN07617.1 PYGN01001214 PSN36624.1 KQ414617 KOC68377.1 KQ981993 KYN30987.1 ADTU01026048 ADTU01026049 CH963848 EDW73915.2 KQ976543 KYM81019.1 AXCN02000497 APCN01001489 KQ982562 KYQ54846.1 GDRN01054278 JAI66101.1 KA645403 AFP60032.1 GBXI01008943 GBXI01003541 JAD05349.1 JAD10751.1 CH954179 EDV57029.1 JXJN01009092 CM000158 EDW92562.1 KRK00579.1 GAMC01013491 JAB93064.1 GFDL01001453 JAV33592.1 AJVK01025461 AJVK01025462 CM000362 CM002911 EDX08575.1 KMY96376.1 CCAG010014920 AY455866 AE013599 AY102683 AAM27512.1 AAR19767.1 CH480838 EDW49216.1 CH940648 EDW61557.1 KRF80062.1 KRF80063.1 GFDL01001513 JAV33532.1
Proteomes
UP000007266
UP000235965
UP000027135
UP000000305
UP000076858
UP000002358
+ More
UP000008237 UP000076502 UP000000311 UP000215335 UP000279307 UP000053097 UP000037069 UP000008744 UP000001819 UP000268350 UP000092553 UP000078200 UP000078492 UP000078542 UP000095300 UP000245037 UP000053825 UP000078541 UP000005205 UP000007798 UP000078540 UP000075886 UP000005203 UP000192221 UP000076407 UP000075840 UP000091820 UP000075809 UP000095301 UP000008711 UP000092460 UP000002282 UP000075884 UP000076408 UP000092462 UP000000304 UP000092444 UP000000803 UP000092443 UP000001292 UP000008792
UP000008237 UP000076502 UP000000311 UP000215335 UP000279307 UP000053097 UP000037069 UP000008744 UP000001819 UP000268350 UP000092553 UP000078200 UP000078492 UP000078542 UP000095300 UP000245037 UP000053825 UP000078541 UP000005205 UP000007798 UP000078540 UP000075886 UP000005203 UP000192221 UP000076407 UP000075840 UP000091820 UP000075809 UP000095301 UP000008711 UP000092460 UP000002282 UP000075884 UP000076408 UP000092462 UP000000304 UP000092444 UP000000803 UP000092443 UP000001292 UP000008792
Pfam
Interpro
IPR002913
START_lipid-bd_dom
+ More
IPR023393 START-like_dom_sf
IPR019498 MENTAL
IPR000799 StAR-like
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR036250 AcylCo_DH-like_C
IPR037069 AcylCoA_DH/ox_N_sf
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009075 AcylCo_DH/oxidase_C
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR002130 Cyclophilin-type_PPIase_dom
IPR001680 WD40_repeat
IPR029000 Cyclophilin-like_dom_sf
IPR023393 START-like_dom_sf
IPR019498 MENTAL
IPR000799 StAR-like
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR036250 AcylCo_DH-like_C
IPR037069 AcylCoA_DH/ox_N_sf
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009075 AcylCo_DH/oxidase_C
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR002130 Cyclophilin-type_PPIase_dom
IPR001680 WD40_repeat
IPR029000 Cyclophilin-like_dom_sf
Gene 3D
ProteinModelPortal
Q3V5Y6
D6WT76
A0A2J7RRK7
A0A067RND7
A0A2J7RRL2
A0A2J7RRI0
+ More
A0A2J7RRJ7 E9FUI1 A0A1Y1N158 A0A1Y1MZS7 A0A0P5V2X9 A0A0P5JA07 A0A0P5LQT0 A0A0P6C0B4 A0A0P5B236 A0A0P5CZM4 A0A0N7ZPD8 A0A0P4YZF7 A0A0P5V270 A0A0P4ZIB0 A0A0P5BKX0 V5GWI2 K7INF7 E2BKV3 A0A154PI58 A0A0N8EK28 E2AXF6 A0A232ENV8 A0A0N8CZL7 A0A0C9RE63 A0A189XGA0 A0A3L8DCB3 A0A026WKG3 A0A0L0CLS7 A0A1L8DY45 A0A1L8DYJ2 B4H561 B5E1A0 A0A3B0J3D2 A0A0K8W3V8 A0A0C9R542 A0A0K8UUC4 A0A034VJP0 A0A0M4EB96 A0A3B0JM71 A0A1A9VWE5 A0A195EET5 A0A195D402 A0A1I8PDW8 A0A2P8XXI2 A0A0L7RBS2 A0A195ESU6 A0A158NTM5 B4MP76 A0A151I2E5 A0A182Q372 A0A087ZMS2 A0A1W4W0I6 A0A182WWT2 A0A182HWG1 A0A1A9WA31 A0A151X343 A0A0P4WBR8 T1PBR4 A0A1I8NFT8 A0A0A1XIE6 B3NQU6 A0A1B0B6G6 B4PBD0 A0A182NJQ0 W8BVC6 A0A182Y264 A0A1Q3G1B7 A0A1B0D5P1 B4QC20 A0A1B0FN74 Q9W145-2 A0A1A9YDA3 B4IH40 Q9W145 B4LJI9 A0A1Q3G164
A0A2J7RRJ7 E9FUI1 A0A1Y1N158 A0A1Y1MZS7 A0A0P5V2X9 A0A0P5JA07 A0A0P5LQT0 A0A0P6C0B4 A0A0P5B236 A0A0P5CZM4 A0A0N7ZPD8 A0A0P4YZF7 A0A0P5V270 A0A0P4ZIB0 A0A0P5BKX0 V5GWI2 K7INF7 E2BKV3 A0A154PI58 A0A0N8EK28 E2AXF6 A0A232ENV8 A0A0N8CZL7 A0A0C9RE63 A0A189XGA0 A0A3L8DCB3 A0A026WKG3 A0A0L0CLS7 A0A1L8DY45 A0A1L8DYJ2 B4H561 B5E1A0 A0A3B0J3D2 A0A0K8W3V8 A0A0C9R542 A0A0K8UUC4 A0A034VJP0 A0A0M4EB96 A0A3B0JM71 A0A1A9VWE5 A0A195EET5 A0A195D402 A0A1I8PDW8 A0A2P8XXI2 A0A0L7RBS2 A0A195ESU6 A0A158NTM5 B4MP76 A0A151I2E5 A0A182Q372 A0A087ZMS2 A0A1W4W0I6 A0A182WWT2 A0A182HWG1 A0A1A9WA31 A0A151X343 A0A0P4WBR8 T1PBR4 A0A1I8NFT8 A0A0A1XIE6 B3NQU6 A0A1B0B6G6 B4PBD0 A0A182NJQ0 W8BVC6 A0A182Y264 A0A1Q3G1B7 A0A1B0D5P1 B4QC20 A0A1B0FN74 Q9W145-2 A0A1A9YDA3 B4IH40 Q9W145 B4LJI9 A0A1Q3G164
PDB
5I9J
E-value=1.07824e-23,
Score=273
Ontologies
GO
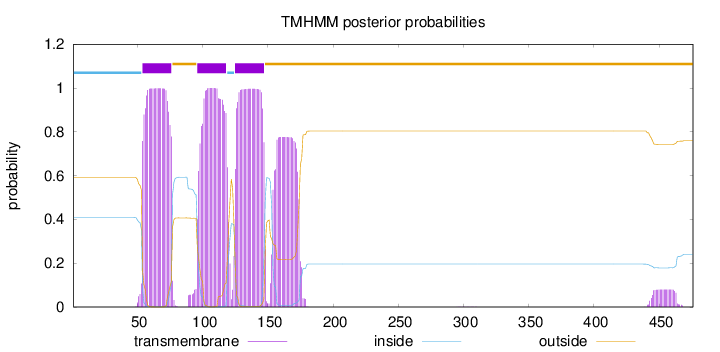
Topology
Subcellular location
Membrane
Length:
476
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
85.15067
Exp number, first 60 AAs:
6.73186
Total prob of N-in:
0.40847
inside
1 - 53
TMhelix
54 - 76
outside
77 - 95
TMhelix
96 - 118
inside
119 - 124
TMhelix
125 - 147
outside
148 - 476
Population Genetic Test Statistics
Pi
272.874736
Theta
174.349428
Tajima's D
3.145123
CLR
0.300752
CSRT
0.987000649967502
Interpretation
Possibly Positive selection
