Gene
KWMTBOMO00917
Pre Gene Modal
BGIBMGA007327
Annotation
UDP-glycosyltransferase_UGT33R1_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
Cytoplasmic Reliability : 1.643 Mitochondrial Reliability : 1.555
Sequence
CDS
ATGAGCTTCGGCACAAACGTTCCTCCTTCGAAGCTGCCTCGTCAGCTGACGCGGATGTTCGCGAGCGTGTTCCGTGAGCTGCCGTACAAGGTGCTGTGGAAGTGGGATCTCGACGTGGTGGAGGGGATGCCGGAGAACGTGAGGACCGGGCGATGGTTCCCGCAGGCTGATGTCTTCAGACATCCGAACGTGAAGCTGGTGATAACCCAGGGAGGGCTGCAGACGTCAGAAGAGGCTATAGAATGCGGGAAACCTCTGATCGGGATACCATTCCTGGCTGACCAATGGCTGAACGTGGACAATTACGTTCACCACGGCATGGGGGTGTATCTGGATGCGGAAACCGTTACTGCTGAGGAGTTCAAAGCGGCCATTGTGGAAGTCATAGAGAATGATAAGTTGGTGAATTGTTAA
Protein
MSFGTNVPPSKLPRQLTRMFASVFRELPYKVLWKWDLDVVEGMPENVRTGRWFPQADVFRHPNVKLVITQGGLQTSEEAIECGKPLIGIPFLADQWLNVDNYVHHGMGVYLDAETVTAEEFKAAIVEVIENDKLVNC
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPT3
H9JCT0
G9LPT4
A0A3B0EEY0
A0A2W1BEA5
G9LPN2
+ More
A0A2W1B8D3 A0A2H1W7S2 A0A2W1B3V4 G9LPN6 A0A191T2F8 A0A212F6V6 A0A0N1II62 A0A024AG00 A0A2W1BEB8 A0A2W1BL20 G9LPP3 A0A2W1BE55 A0A191T1K8 M4M1W9 G9LPN9 A0A2W1BCY8 G9LPP0 A0A212F6X6 A0A2W1B475 A0A0N1IQ70 A0A0N0PE43 A0A191T2J2 A0A2H4WB73 H9JWE5 A0A2H1W0F9 A0A1L8D6T8 A0A191T1L3 A0A0L7KPR9 A0A0L7LGN7 A0A0N1IK49 G9LPS3 G9LPP5 G9LPP1 A0A2I6RJL8 H9JWB7 G9LPN7 A0A2A4JXE4 G9LPS5 A0A2H1VU94 A0A2W1BHP8 A0A2A4IVU8 A0A2W1B3X2 A0A0N1IIW5 A0A2H1WZR7 A0A024AH14 A0A2W1B485 G9LPN3 A0A024AGL3 A0A0N1I2R8 H9JWB6 A0A2H1WTF5 Q1HPX7 A0A0N0P9C8 G9LPS4 A0A191T1L2 H9JWB9 G9LPT1 A0A2W1B9W6 G9LPN5 A0A140EA35 A0A2W1B8G2 G9LPN8 B7TZA3 A0A2W1BQP4 A0A191T2J5 G9LPS9 G9LPS6 F1DG68 A0A191T1L8 G9LPP7 A0A0N1PG92 G9LPS2 A0A0N1PE65 A0A212F6V8 A0A0N1IE25 A0A2W1BBH1 A0A0N1IB27 A0A2H1WSA7 A0A2W1BAD4 H9JWC2 H9JWE6 G9LPP6 A0A2H4WB75 D2JLK8 A0A2W1BA70 G9LPS7 A0A0N0PEI5 A0A212FKM1 G9LPS8 A0A2H1VUS4 A0A2R4FXG7 A0A1W4XIA4 A0A0N1ICS6 A0A1W4XHT9
A0A2W1B8D3 A0A2H1W7S2 A0A2W1B3V4 G9LPN6 A0A191T2F8 A0A212F6V6 A0A0N1II62 A0A024AG00 A0A2W1BEB8 A0A2W1BL20 G9LPP3 A0A2W1BE55 A0A191T1K8 M4M1W9 G9LPN9 A0A2W1BCY8 G9LPP0 A0A212F6X6 A0A2W1B475 A0A0N1IQ70 A0A0N0PE43 A0A191T2J2 A0A2H4WB73 H9JWE5 A0A2H1W0F9 A0A1L8D6T8 A0A191T1L3 A0A0L7KPR9 A0A0L7LGN7 A0A0N1IK49 G9LPS3 G9LPP5 G9LPP1 A0A2I6RJL8 H9JWB7 G9LPN7 A0A2A4JXE4 G9LPS5 A0A2H1VU94 A0A2W1BHP8 A0A2A4IVU8 A0A2W1B3X2 A0A0N1IIW5 A0A2H1WZR7 A0A024AH14 A0A2W1B485 G9LPN3 A0A024AGL3 A0A0N1I2R8 H9JWB6 A0A2H1WTF5 Q1HPX7 A0A0N0P9C8 G9LPS4 A0A191T1L2 H9JWB9 G9LPT1 A0A2W1B9W6 G9LPN5 A0A140EA35 A0A2W1B8G2 G9LPN8 B7TZA3 A0A2W1BQP4 A0A191T2J5 G9LPS9 G9LPS6 F1DG68 A0A191T1L8 G9LPP7 A0A0N1PG92 G9LPS2 A0A0N1PE65 A0A212F6V8 A0A0N1IE25 A0A2W1BBH1 A0A0N1IB27 A0A2H1WSA7 A0A2W1BAD4 H9JWC2 H9JWE6 G9LPP6 A0A2H4WB75 D2JLK8 A0A2W1BA70 G9LPS7 A0A0N0PEI5 A0A212FKM1 G9LPS8 A0A2H1VUS4 A0A2R4FXG7 A0A1W4XIA4 A0A0N1ICS6 A0A1W4XHT9
EC Number
2.4.1.17
Pubmed
EMBL
JQ070240
AEW43156.1
BABH01040891
JQ070241
AEW43157.1
RBVL01000113
+ More
RKO12004.1 KZ150360 PZC71190.1 JQ070189 AEW43105.1 PZC71191.1 ODYU01006890 SOQ49139.1 PZC71192.1 JQ070193 AEW43109.1 KU680280 ANI21986.1 AGBW02009962 OWR49472.1 KQ459937 KPJ19199.1 KF777109 AHY99680.1 PZC71200.1 KZ150193 PZC72373.1 JQ070200 AEW43116.1 PZC71196.1 KU680282 ANI21988.1 JQ747500 AGG36457.1 JQ070196 AEW43112.1 PZC71197.1 JQ070197 AEW43113.1 OWR49474.1 PZC71189.1 KPJ19197.1 KPJ19195.1 KU680284 ANI21990.1 KY202933 AUC64277.1 BABH01029271 BABH01029272 ODYU01005543 SOQ46493.1 GEYN01000079 JAV02050.1 KU680288 ANI21994.1 JTDY01007346 KOB65277.1 JTDY01001195 KOB74580.1 KQ459680 KPJ20475.1 JQ070230 AEW43146.1 JQ070202 AEW43118.1 JQ070198 AEW43114.1 KY304474 AUN86405.1 BABH01029269 BABH01029270 JQ070194 AEW43110.1 NWSH01000435 PCG76446.1 JQ070232 AEW43148.1 ODYU01004485 SOQ44409.1 PZC71193.1 NWSH01005967 PCG63789.1 PZC71202.1 KPJ20476.1 ODYU01011876 SOQ57874.1 KF777111 AHY99682.1 PZC71199.1 JQ070190 AEW43106.1 KF777112 AHY99683.1 KQ459547 KPJ00048.1 ODYU01010933 SOQ56350.1 DQ443275 ABF51364.1 KPJ00042.1 JQ070231 AEW43147.1 KU680287 ANI21993.1 BABH01029259 BABH01029260 JQ070238 AEW43154.1 PZC72369.1 JQ070192 AEW43108.1 KU214506 AMK97472.1 PZC71201.1 JQ070195 AEW43111.1 FJ418685 ACJ48963.1 KZ149974 PZC75964.1 KU680289 ANI21995.1 JQ070236 AEW43152.1 JQ070233 AEW43149.1 HQ833816 ADY17534.1 KU680281 ANI21987.1 JQ070204 AEW43120.1 KQ460636 KPJ13398.1 JQ070229 AEW43145.1 KPJ00045.1 OWR49475.1 KPJ13397.1 PZC71194.1 KPJ00043.1 ODYU01010294 SOQ55304.1 PZC72372.1 BABH01029238 BABH01029239 BABH01029240 BABH01029273 BABH01029274 JQ070203 AEW43119.1 KY202945 AUC64289.1 GQ915324 ACZ97418.2 PZC71195.1 JQ070234 AEW43150.1 KPJ19193.1 AGBW02008024 OWR54293.1 JQ070235 AEW43151.1 ODYU01004546 SOQ44548.1 MF034857 AVT42223.1 KPJ00041.1
RKO12004.1 KZ150360 PZC71190.1 JQ070189 AEW43105.1 PZC71191.1 ODYU01006890 SOQ49139.1 PZC71192.1 JQ070193 AEW43109.1 KU680280 ANI21986.1 AGBW02009962 OWR49472.1 KQ459937 KPJ19199.1 KF777109 AHY99680.1 PZC71200.1 KZ150193 PZC72373.1 JQ070200 AEW43116.1 PZC71196.1 KU680282 ANI21988.1 JQ747500 AGG36457.1 JQ070196 AEW43112.1 PZC71197.1 JQ070197 AEW43113.1 OWR49474.1 PZC71189.1 KPJ19197.1 KPJ19195.1 KU680284 ANI21990.1 KY202933 AUC64277.1 BABH01029271 BABH01029272 ODYU01005543 SOQ46493.1 GEYN01000079 JAV02050.1 KU680288 ANI21994.1 JTDY01007346 KOB65277.1 JTDY01001195 KOB74580.1 KQ459680 KPJ20475.1 JQ070230 AEW43146.1 JQ070202 AEW43118.1 JQ070198 AEW43114.1 KY304474 AUN86405.1 BABH01029269 BABH01029270 JQ070194 AEW43110.1 NWSH01000435 PCG76446.1 JQ070232 AEW43148.1 ODYU01004485 SOQ44409.1 PZC71193.1 NWSH01005967 PCG63789.1 PZC71202.1 KPJ20476.1 ODYU01011876 SOQ57874.1 KF777111 AHY99682.1 PZC71199.1 JQ070190 AEW43106.1 KF777112 AHY99683.1 KQ459547 KPJ00048.1 ODYU01010933 SOQ56350.1 DQ443275 ABF51364.1 KPJ00042.1 JQ070231 AEW43147.1 KU680287 ANI21993.1 BABH01029259 BABH01029260 JQ070238 AEW43154.1 PZC72369.1 JQ070192 AEW43108.1 KU214506 AMK97472.1 PZC71201.1 JQ070195 AEW43111.1 FJ418685 ACJ48963.1 KZ149974 PZC75964.1 KU680289 ANI21995.1 JQ070236 AEW43152.1 JQ070233 AEW43149.1 HQ833816 ADY17534.1 KU680281 ANI21987.1 JQ070204 AEW43120.1 KQ460636 KPJ13398.1 JQ070229 AEW43145.1 KPJ00045.1 OWR49475.1 KPJ13397.1 PZC71194.1 KPJ00043.1 ODYU01010294 SOQ55304.1 PZC72372.1 BABH01029238 BABH01029239 BABH01029240 BABH01029273 BABH01029274 JQ070203 AEW43119.1 KY202945 AUC64289.1 GQ915324 ACZ97418.2 PZC71195.1 JQ070234 AEW43150.1 KPJ19193.1 AGBW02008024 OWR54293.1 JQ070235 AEW43151.1 ODYU01004546 SOQ44548.1 MF034857 AVT42223.1 KPJ00041.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
G9LPT3
H9JCT0
G9LPT4
A0A3B0EEY0
A0A2W1BEA5
G9LPN2
+ More
A0A2W1B8D3 A0A2H1W7S2 A0A2W1B3V4 G9LPN6 A0A191T2F8 A0A212F6V6 A0A0N1II62 A0A024AG00 A0A2W1BEB8 A0A2W1BL20 G9LPP3 A0A2W1BE55 A0A191T1K8 M4M1W9 G9LPN9 A0A2W1BCY8 G9LPP0 A0A212F6X6 A0A2W1B475 A0A0N1IQ70 A0A0N0PE43 A0A191T2J2 A0A2H4WB73 H9JWE5 A0A2H1W0F9 A0A1L8D6T8 A0A191T1L3 A0A0L7KPR9 A0A0L7LGN7 A0A0N1IK49 G9LPS3 G9LPP5 G9LPP1 A0A2I6RJL8 H9JWB7 G9LPN7 A0A2A4JXE4 G9LPS5 A0A2H1VU94 A0A2W1BHP8 A0A2A4IVU8 A0A2W1B3X2 A0A0N1IIW5 A0A2H1WZR7 A0A024AH14 A0A2W1B485 G9LPN3 A0A024AGL3 A0A0N1I2R8 H9JWB6 A0A2H1WTF5 Q1HPX7 A0A0N0P9C8 G9LPS4 A0A191T1L2 H9JWB9 G9LPT1 A0A2W1B9W6 G9LPN5 A0A140EA35 A0A2W1B8G2 G9LPN8 B7TZA3 A0A2W1BQP4 A0A191T2J5 G9LPS9 G9LPS6 F1DG68 A0A191T1L8 G9LPP7 A0A0N1PG92 G9LPS2 A0A0N1PE65 A0A212F6V8 A0A0N1IE25 A0A2W1BBH1 A0A0N1IB27 A0A2H1WSA7 A0A2W1BAD4 H9JWC2 H9JWE6 G9LPP6 A0A2H4WB75 D2JLK8 A0A2W1BA70 G9LPS7 A0A0N0PEI5 A0A212FKM1 G9LPS8 A0A2H1VUS4 A0A2R4FXG7 A0A1W4XIA4 A0A0N1ICS6 A0A1W4XHT9
A0A2W1B8D3 A0A2H1W7S2 A0A2W1B3V4 G9LPN6 A0A191T2F8 A0A212F6V6 A0A0N1II62 A0A024AG00 A0A2W1BEB8 A0A2W1BL20 G9LPP3 A0A2W1BE55 A0A191T1K8 M4M1W9 G9LPN9 A0A2W1BCY8 G9LPP0 A0A212F6X6 A0A2W1B475 A0A0N1IQ70 A0A0N0PE43 A0A191T2J2 A0A2H4WB73 H9JWE5 A0A2H1W0F9 A0A1L8D6T8 A0A191T1L3 A0A0L7KPR9 A0A0L7LGN7 A0A0N1IK49 G9LPS3 G9LPP5 G9LPP1 A0A2I6RJL8 H9JWB7 G9LPN7 A0A2A4JXE4 G9LPS5 A0A2H1VU94 A0A2W1BHP8 A0A2A4IVU8 A0A2W1B3X2 A0A0N1IIW5 A0A2H1WZR7 A0A024AH14 A0A2W1B485 G9LPN3 A0A024AGL3 A0A0N1I2R8 H9JWB6 A0A2H1WTF5 Q1HPX7 A0A0N0P9C8 G9LPS4 A0A191T1L2 H9JWB9 G9LPT1 A0A2W1B9W6 G9LPN5 A0A140EA35 A0A2W1B8G2 G9LPN8 B7TZA3 A0A2W1BQP4 A0A191T2J5 G9LPS9 G9LPS6 F1DG68 A0A191T1L8 G9LPP7 A0A0N1PG92 G9LPS2 A0A0N1PE65 A0A212F6V8 A0A0N1IE25 A0A2W1BBH1 A0A0N1IB27 A0A2H1WSA7 A0A2W1BAD4 H9JWC2 H9JWE6 G9LPP6 A0A2H4WB75 D2JLK8 A0A2W1BA70 G9LPS7 A0A0N0PEI5 A0A212FKM1 G9LPS8 A0A2H1VUS4 A0A2R4FXG7 A0A1W4XIA4 A0A0N1ICS6 A0A1W4XHT9
PDB
2O6L
E-value=1.43049e-15,
Score=195
Ontologies
GO
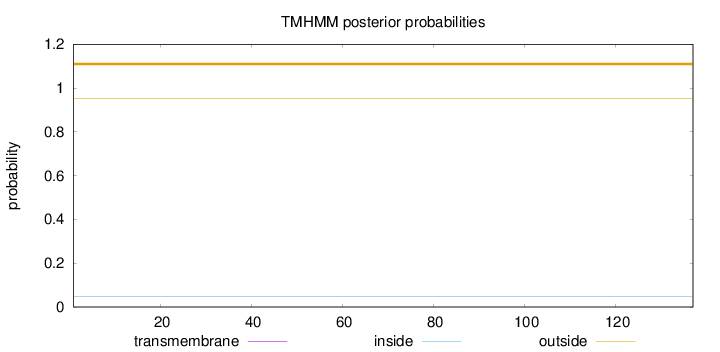
Topology
Subcellular location
Membrane
Length:
137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00053
Exp number, first 60 AAs:
0.00053
Total prob of N-in:
0.04746
outside
1 - 137
Population Genetic Test Statistics
Pi
268.619942
Theta
209.63158
Tajima's D
0.90139
CLR
0.369222
CSRT
0.632018399080046
Interpretation
Uncertain
