Gene
KWMTBOMO00913
Pre Gene Modal
BGIBMGA009650
Annotation
PREDICTED:_mothers_against_decapentaplegic_homolog_3_[Papilio_polytes]
Full name
Mothers against decapentaplegic homolog
+ More
Mothers against decapentaplegic homolog 3
Mothers against decapentaplegic homolog 3
Alternative Name
SMAD family member
SMAD family member 3
SMAD family member 3
Transcription factor
Location in the cell
Nuclear Reliability : 2.556
Sequence
CDS
ATGTTCCCGTTAACTCCGCCGGTCGTGAAGCGATTGTTGGGCTGGAAAAAAGGACCCGAGGGATCGTCGGCCGCAGAAGATAAATGGTCCGAGAAGGCCGTAAAGAGTTTGGTAAAAAAATTAAAGAAAAGCGGGGCTTTAGAAGAGTTAGAAAGGGCAATCTCGAGTCAGAACAGTCATACGAAATGCGTTACGATACCCAGGGTGAAGCCGAATGAGAGTGGCATCAACGGACAGTACAGGAAAGGCCTACCGCATGTGGTTTACTGCAGACTCTGGCGTTGGCCTCAGTTACAGAGCCAACATGAACTGAAGCCAGTGGACCATTGCGAGTATGCATACCAGCTGAAGAAGGACGAGGTGTGCATTAATCCCTATCATTACAACAAAATCGATTCGCCTGCTCTTCCCCCGATCCTGGTGCGTCGCTGTAGCGAGGGCGAGGTCCGCGCCCCTCCGCCCTACGCCGACTACCACCAGCCAATACTGCACGAGCACCCTGACGTTGCCATGCAGTCTGGTACAGGACACAGCGCACTGTATCTCGAGGCTACACTTGCACAACACGTGCCGGGCAACACAACTGTACACGTGAGCTCCTCAACAGTGGAGACTCCGCCCCCAGGCTACATCAGCGAAGATGGAGACCCGATGGACCACAATGACAACATGAACCTCACACGTCTAAGTCCGTCACCTGGCGGCCTGGCGTCCGAGGCCTCGCCGGTGCTGTACCACGAGCCGGCGTTCTGGTGCAGCATCAGCTACTACGAGCTGAACACCAGGGTCGGGGAGACGTTCCACGCCTCGCAACCTTCCATCACGGTCGACGGATTCACCGACCCGAGCAATAGTGAGAGATTTTGTCTCGGCCTGCTTTCGAACGTGAACAGGAACGAAGTCGTCGAGCAAACGCGCAGACACATCGGGAAAGGAGTCCGGCTGTATTATATAGGGGGTGAGGTCTTCGCGGAGTGCCTGAGCGACTCCTCGATATTCGTGCAGAGTCCGAACTGCAACCAGCGCTATGGCTGGCATCCGGCCACCGTTTGTAAAATACCACCAGGTTGCAACTTGAAGATATTTAATAATCAAGAATTCGCGGCCCTCCTGTCGCAGTCGGTCTCGCAGGGGTTCGAAGCCGTCTTCCAGCTGACCAGGATGTGCACAATACGAATGAGCTTCGTCAAGGGGTGGGGGGCTGAATACAGACGGCAAACTGTAACTTCAACTCCGTGTTGGATCGAGCTGCATCTCAACGGTCCTCTGCAGTGGCTGGACAGAGTCCTCACACAGATGGGCTCTCCGCGTCTGCCGTGCTCCTCTATGTCCTAG
Protein
MFPLTPPVVKRLLGWKKGPEGSSAAEDKWSEKAVKSLVKKLKKSGALEELERAISSQNSHTKCVTIPRVKPNESGINGQYRKGLPHVVYCRLWRWPQLQSQHELKPVDHCEYAYQLKKDEVCINPYHYNKIDSPALPPILVRRCSEGEVRAPPPYADYHQPILHEHPDVAMQSGTGHSALYLEATLAQHVPGNTTVHVSSSTVETPPPGYISEDGDPMDHNDNMNLTRLSPSPGGLASEASPVLYHEPAFWCSISYYELNTRVGETFHASQPSITVDGFTDPSNSERFCLGLLSNVNRNEVVEQTRRHIGKGVRLYYIGGEVFAECLSDSSIFVQSPNCNQRYGWHPATVCKIPPGCNLKIFNNQEFAALLSQSVSQGFEAVFQLTRMCTIRMSFVKGWGAEYRRQTVTSTPCWIELHLNGPLQWLDRVLTQMGSPRLPCSSMS
Summary
Description
Transcriptional modulator activated by TGF-beta (transforming growth factor) and activin type 1 receptor kinase. SMAD3 is a receptor-regulated SMAD (R-SMAD) (By similarity).
Subunit
Interacts with SARA (SMAD anchor for receptor activation); form trimers with another SMAD3 and the co-SMAD SMAD4.
Similarity
Belongs to the dwarfin/SMAD family.
Keywords
Complete proteome
Cytoplasm
DNA-binding
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Zinc
Feature
chain Mothers against decapentaplegic homolog 3
Uniprot
H9JJE9
A0A220QT60
A0A2H1VIR2
A0A194PTI1
A0A212F8N9
A0A0N1IH69
+ More
U5EXD3 A0A1L8DMQ0 F8V242 A0A067QRP6 K7IP92 A0A195ET96 A0A158NVW1 A0A336LKU2 A0A0K8TML2 A0A0L7RBH0 A0A1E1XLS3 A0A1E1XFN2 A0A131Z076 A0A131XW48 E9G7J0 A0A131YYC5 A0A1B6KEL6 A0A2R5LI54 A0A1B6GH93 A0A087T8C0 A0A3B5KT50 A0A3B3XI70 M3ZMX8 J7FXQ7 A0A315W4G7 A0A087XSS2 A0A3B3DUI3 A0A3B3TZU3 A0A3P9P210 A0A3B5BG60 C3Y6V2 A0A142EFR7 A0A0F8CFE8 A0A3B4YLL3 A0A3B4TX57 A0A3P8TX98 A0A091EAG3 A0A0F7ZD79 A0A3P8PYH6 A0A3P8VEX9 A0A3B3CNI8 H2MLR1 A0A3P9J8K3 A0A3P8VN55 A0A3B5AWN7 I3KEB1 Q8AY15 A0A0P7V3D6 A0A1S3K8S6 A0A1V4JDJ5 A0A0Q3PU66 P84023 A0A3B4XS14 A0A3B4TGC7 A0A146Z4J3 A0A091GFE6 F6S0M0 A0A218V0C7 U3KEJ5 A0A3P9ILG9 H2LR51 A0A1A8LN14 A0A1A8VDT5 A0A1A8QAR2 A0A091TN31 A0A1A7X9E8 A0A1A7Z463 B2XS64 A0A1A8IPB1 A0A093JC11 A0A1S3K891 G3PJ40 A0A3P8VEY6 A0A3B4ZAV1 A0A384C4Z0 A0A1L8DLE1 A0A099YXV9 A0A091GCW3 A0A1U7TYB8 A0A2I4BN26 A0A091SZL2 A0A091PJT1 E0VAB2 A0A3B5KD16 H2UPJ8 A0A096MCX0 Q68EP5 A2PZA6 A0A1A8DS60 A0A1A8UG63 F6UYG1 Q90YE5 A0A1L8GSA7 A0A1A8MZ22
U5EXD3 A0A1L8DMQ0 F8V242 A0A067QRP6 K7IP92 A0A195ET96 A0A158NVW1 A0A336LKU2 A0A0K8TML2 A0A0L7RBH0 A0A1E1XLS3 A0A1E1XFN2 A0A131Z076 A0A131XW48 E9G7J0 A0A131YYC5 A0A1B6KEL6 A0A2R5LI54 A0A1B6GH93 A0A087T8C0 A0A3B5KT50 A0A3B3XI70 M3ZMX8 J7FXQ7 A0A315W4G7 A0A087XSS2 A0A3B3DUI3 A0A3B3TZU3 A0A3P9P210 A0A3B5BG60 C3Y6V2 A0A142EFR7 A0A0F8CFE8 A0A3B4YLL3 A0A3B4TX57 A0A3P8TX98 A0A091EAG3 A0A0F7ZD79 A0A3P8PYH6 A0A3P8VEX9 A0A3B3CNI8 H2MLR1 A0A3P9J8K3 A0A3P8VN55 A0A3B5AWN7 I3KEB1 Q8AY15 A0A0P7V3D6 A0A1S3K8S6 A0A1V4JDJ5 A0A0Q3PU66 P84023 A0A3B4XS14 A0A3B4TGC7 A0A146Z4J3 A0A091GFE6 F6S0M0 A0A218V0C7 U3KEJ5 A0A3P9ILG9 H2LR51 A0A1A8LN14 A0A1A8VDT5 A0A1A8QAR2 A0A091TN31 A0A1A7X9E8 A0A1A7Z463 B2XS64 A0A1A8IPB1 A0A093JC11 A0A1S3K891 G3PJ40 A0A3P8VEY6 A0A3B4ZAV1 A0A384C4Z0 A0A1L8DLE1 A0A099YXV9 A0A091GCW3 A0A1U7TYB8 A0A2I4BN26 A0A091SZL2 A0A091PJT1 E0VAB2 A0A3B5KD16 H2UPJ8 A0A096MCX0 Q68EP5 A2PZA6 A0A1A8DS60 A0A1A8UG63 F6UYG1 Q90YE5 A0A1L8GSA7 A0A1A8MZ22
Pubmed
EMBL
BABH01017772
BABH01017773
KY328719
ASK12087.1
ODYU01002789
SOQ40717.1
+ More
KQ459593 KPI96622.1 AGBW02009700 OWR50091.1 KQ460280 KPJ16324.1 GANO01000124 JAB59747.1 GFDF01006440 JAV07644.1 JN006979 AEI25997.1 KK853098 KDR11392.1 KQ981993 KYN31104.1 ADTU01027543 ADTU01027544 ADTU01027545 ADTU01027546 ADTU01027547 ADTU01027548 UFQT01000004 SSX17279.1 GDAI01002195 JAI15408.1 KQ414617 KOC68091.1 GFAA01003398 JAU00037.1 GFAC01001108 JAT98080.1 GEDV01004375 JAP84182.1 GEFM01004283 GEGO01001769 JAP71513.1 JAR93635.1 GL732534 EFX84472.1 GEDV01004374 JAP84183.1 GEBQ01030077 JAT09900.1 GGLE01004969 MBY09095.1 GECZ01007957 JAS61812.1 KK113918 KFM61359.1 JX103494 AFP57672.1 NHOQ01000384 PWA30499.1 AYCK01006720 GG666488 EEN64062.1 KT900137 AMQ47456.1 KQ042192 KKF18150.1 KK717966 KFO53469.1 GBEX01001891 JAI12669.1 AERX01005350 CR352307 CR927368 BC129150 AY134492 AAI29151.1 AAN08606.1 JARO02004087 KPP69180.1 LSYS01007908 OPJ70323.1 LMAW01000980 KQK84747.1 AY391265 GCES01025591 JAR60732.1 KL448190 KFO81115.1 MUZQ01000079 OWK59419.1 AGTO01000581 HAEF01008251 SBR46235.1 HAEJ01017569 SBS58026.1 HAEH01010710 SBR90407.1 KK457768 KFQ77398.1 HADW01013034 SBP14434.1 HADW01004168 HADX01015072 SBP37304.1 EU137731 ABX57736.1 HAED01012719 SBQ99117.1 KL205903 KFV76434.1 GFDF01006822 JAV07262.1 KL887587 KGL74979.1 KL448063 KFO79778.1 KK491972 KFQ63773.1 KK675067 KFQ08157.1 DS235005 EEB10318.1 AYCK01014919 BC080156 AAH80156.1 AB267397 BAF45857.1 HAEA01007174 SBQ35654.1 HADY01018552 HAEJ01005666 SBS46123.1 BC169420 BC170316 AJ311059 CM004470 AAI69420.1 CAC38118.1 OCT89841.1 CM004471 OCT86733.1 HAEF01020939 HAEG01013589 SBR62098.1
KQ459593 KPI96622.1 AGBW02009700 OWR50091.1 KQ460280 KPJ16324.1 GANO01000124 JAB59747.1 GFDF01006440 JAV07644.1 JN006979 AEI25997.1 KK853098 KDR11392.1 KQ981993 KYN31104.1 ADTU01027543 ADTU01027544 ADTU01027545 ADTU01027546 ADTU01027547 ADTU01027548 UFQT01000004 SSX17279.1 GDAI01002195 JAI15408.1 KQ414617 KOC68091.1 GFAA01003398 JAU00037.1 GFAC01001108 JAT98080.1 GEDV01004375 JAP84182.1 GEFM01004283 GEGO01001769 JAP71513.1 JAR93635.1 GL732534 EFX84472.1 GEDV01004374 JAP84183.1 GEBQ01030077 JAT09900.1 GGLE01004969 MBY09095.1 GECZ01007957 JAS61812.1 KK113918 KFM61359.1 JX103494 AFP57672.1 NHOQ01000384 PWA30499.1 AYCK01006720 GG666488 EEN64062.1 KT900137 AMQ47456.1 KQ042192 KKF18150.1 KK717966 KFO53469.1 GBEX01001891 JAI12669.1 AERX01005350 CR352307 CR927368 BC129150 AY134492 AAI29151.1 AAN08606.1 JARO02004087 KPP69180.1 LSYS01007908 OPJ70323.1 LMAW01000980 KQK84747.1 AY391265 GCES01025591 JAR60732.1 KL448190 KFO81115.1 MUZQ01000079 OWK59419.1 AGTO01000581 HAEF01008251 SBR46235.1 HAEJ01017569 SBS58026.1 HAEH01010710 SBR90407.1 KK457768 KFQ77398.1 HADW01013034 SBP14434.1 HADW01004168 HADX01015072 SBP37304.1 EU137731 ABX57736.1 HAED01012719 SBQ99117.1 KL205903 KFV76434.1 GFDF01006822 JAV07262.1 KL887587 KGL74979.1 KL448063 KFO79778.1 KK491972 KFQ63773.1 KK675067 KFQ08157.1 DS235005 EEB10318.1 AYCK01014919 BC080156 AAH80156.1 AB267397 BAF45857.1 HAEA01007174 SBQ35654.1 HADY01018552 HAEJ01005666 SBS46123.1 BC169420 BC170316 AJ311059 CM004470 AAI69420.1 CAC38118.1 OCT89841.1 CM004471 OCT86733.1 HAEF01020939 HAEG01013589 SBR62098.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000027135
UP000002358
+ More
UP000078541 UP000005205 UP000053825 UP000000305 UP000054359 UP000261380 UP000261480 UP000002852 UP000028760 UP000261560 UP000261500 UP000242638 UP000261400 UP000001554 UP000261360 UP000261420 UP000265080 UP000052976 UP000265100 UP000265120 UP000001038 UP000265180 UP000265200 UP000005207 UP000000437 UP000034805 UP000192224 UP000085678 UP000190648 UP000051836 UP000000539 UP000053760 UP000002280 UP000197619 UP000016665 UP000053584 UP000007635 UP000261680 UP000291021 UP000053641 UP000189704 UP000192220 UP000009046 UP000005226 UP000002281 UP000186698
UP000078541 UP000005205 UP000053825 UP000000305 UP000054359 UP000261380 UP000261480 UP000002852 UP000028760 UP000261560 UP000261500 UP000242638 UP000261400 UP000001554 UP000261360 UP000261420 UP000265080 UP000052976 UP000265100 UP000265120 UP000001038 UP000265180 UP000265200 UP000005207 UP000000437 UP000034805 UP000192224 UP000085678 UP000190648 UP000051836 UP000000539 UP000053760 UP000002280 UP000197619 UP000016665 UP000053584 UP000007635 UP000261680 UP000291021 UP000053641 UP000189704 UP000192220 UP000009046 UP000005226 UP000002281 UP000186698
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JJE9
A0A220QT60
A0A2H1VIR2
A0A194PTI1
A0A212F8N9
A0A0N1IH69
+ More
U5EXD3 A0A1L8DMQ0 F8V242 A0A067QRP6 K7IP92 A0A195ET96 A0A158NVW1 A0A336LKU2 A0A0K8TML2 A0A0L7RBH0 A0A1E1XLS3 A0A1E1XFN2 A0A131Z076 A0A131XW48 E9G7J0 A0A131YYC5 A0A1B6KEL6 A0A2R5LI54 A0A1B6GH93 A0A087T8C0 A0A3B5KT50 A0A3B3XI70 M3ZMX8 J7FXQ7 A0A315W4G7 A0A087XSS2 A0A3B3DUI3 A0A3B3TZU3 A0A3P9P210 A0A3B5BG60 C3Y6V2 A0A142EFR7 A0A0F8CFE8 A0A3B4YLL3 A0A3B4TX57 A0A3P8TX98 A0A091EAG3 A0A0F7ZD79 A0A3P8PYH6 A0A3P8VEX9 A0A3B3CNI8 H2MLR1 A0A3P9J8K3 A0A3P8VN55 A0A3B5AWN7 I3KEB1 Q8AY15 A0A0P7V3D6 A0A1S3K8S6 A0A1V4JDJ5 A0A0Q3PU66 P84023 A0A3B4XS14 A0A3B4TGC7 A0A146Z4J3 A0A091GFE6 F6S0M0 A0A218V0C7 U3KEJ5 A0A3P9ILG9 H2LR51 A0A1A8LN14 A0A1A8VDT5 A0A1A8QAR2 A0A091TN31 A0A1A7X9E8 A0A1A7Z463 B2XS64 A0A1A8IPB1 A0A093JC11 A0A1S3K891 G3PJ40 A0A3P8VEY6 A0A3B4ZAV1 A0A384C4Z0 A0A1L8DLE1 A0A099YXV9 A0A091GCW3 A0A1U7TYB8 A0A2I4BN26 A0A091SZL2 A0A091PJT1 E0VAB2 A0A3B5KD16 H2UPJ8 A0A096MCX0 Q68EP5 A2PZA6 A0A1A8DS60 A0A1A8UG63 F6UYG1 Q90YE5 A0A1L8GSA7 A0A1A8MZ22
U5EXD3 A0A1L8DMQ0 F8V242 A0A067QRP6 K7IP92 A0A195ET96 A0A158NVW1 A0A336LKU2 A0A0K8TML2 A0A0L7RBH0 A0A1E1XLS3 A0A1E1XFN2 A0A131Z076 A0A131XW48 E9G7J0 A0A131YYC5 A0A1B6KEL6 A0A2R5LI54 A0A1B6GH93 A0A087T8C0 A0A3B5KT50 A0A3B3XI70 M3ZMX8 J7FXQ7 A0A315W4G7 A0A087XSS2 A0A3B3DUI3 A0A3B3TZU3 A0A3P9P210 A0A3B5BG60 C3Y6V2 A0A142EFR7 A0A0F8CFE8 A0A3B4YLL3 A0A3B4TX57 A0A3P8TX98 A0A091EAG3 A0A0F7ZD79 A0A3P8PYH6 A0A3P8VEX9 A0A3B3CNI8 H2MLR1 A0A3P9J8K3 A0A3P8VN55 A0A3B5AWN7 I3KEB1 Q8AY15 A0A0P7V3D6 A0A1S3K8S6 A0A1V4JDJ5 A0A0Q3PU66 P84023 A0A3B4XS14 A0A3B4TGC7 A0A146Z4J3 A0A091GFE6 F6S0M0 A0A218V0C7 U3KEJ5 A0A3P9ILG9 H2LR51 A0A1A8LN14 A0A1A8VDT5 A0A1A8QAR2 A0A091TN31 A0A1A7X9E8 A0A1A7Z463 B2XS64 A0A1A8IPB1 A0A093JC11 A0A1S3K891 G3PJ40 A0A3P8VEY6 A0A3B4ZAV1 A0A384C4Z0 A0A1L8DLE1 A0A099YXV9 A0A091GCW3 A0A1U7TYB8 A0A2I4BN26 A0A091SZL2 A0A091PJT1 E0VAB2 A0A3B5KD16 H2UPJ8 A0A096MCX0 Q68EP5 A2PZA6 A0A1A8DS60 A0A1A8UG63 F6UYG1 Q90YE5 A0A1L8GSA7 A0A1A8MZ22
PDB
1KHX
E-value=9.8448e-113,
Score=1041
Ontologies
PATHWAY
GO
GO:0005634
GO:0007179
GO:0005737
GO:0003677
GO:0005667
GO:0003700
GO:0007498
GO:0060215
GO:0006355
GO:0043565
GO:0000987
GO:0046872
GO:0071141
GO:0032332
GO:0005886
GO:0002520
GO:0048340
GO:0045429
GO:0071144
GO:0000122
GO:1902895
GO:0048589
GO:0007183
GO:0050678
GO:0008270
GO:0070878
GO:0050776
GO:0030878
GO:0032909
GO:0031490
GO:0030308
GO:0001657
GO:0061045
GO:0042803
GO:0043425
GO:0043235
GO:0046982
GO:0060395
GO:0031962
GO:0048617
GO:0010718
GO:0050728
GO:0019049
GO:0005829
GO:0005160
GO:0097191
GO:0070410
GO:0045216
GO:0030618
GO:0002076
GO:0042110
GO:0019901
GO:0001947
GO:0005654
GO:0043130
GO:0006955
GO:0070412
GO:0048701
GO:0045599
GO:0019902
GO:0009880
GO:1901203
GO:0070306
GO:0033689
GO:0051481
GO:0005518
GO:0001889
GO:0035259
GO:0045668
GO:0031625
GO:0007050
GO:0051098
GO:0001756
GO:0000790
GO:0003712
GO:0001666
GO:0005637
GO:0001707
GO:0006919
GO:0016202
GO:0017015
GO:0045930
GO:0007492
GO:0017151
GO:0001102
GO:0001228
GO:0000978
GO:0001701
GO:0060039
GO:0038092
GO:0005622
GO:0005515
GO:0000166
GO:0004887
GO:0003707
GO:0016021
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
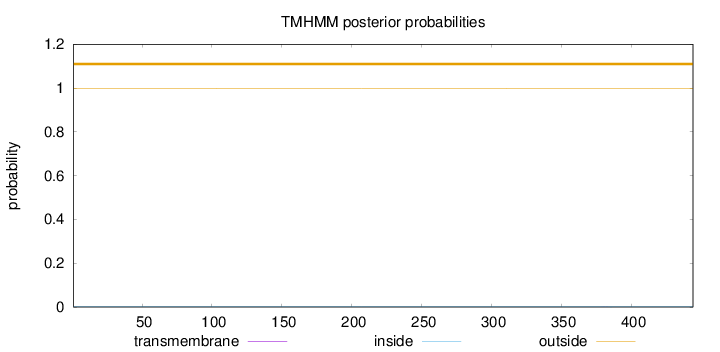
Length:
444
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00379
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00071
outside
1 - 444
Population Genetic Test Statistics
Pi
245.182118
Theta
163.773767
Tajima's D
1.32497
CLR
0.197952
CSRT
0.747112644367782
Interpretation
Uncertain
