Pre Gene Modal
BGIBMGA009779
Annotation
PREDICTED:_unconventional_myosin-Va_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.435
Sequence
CDS
ATGTCGGTCCAGCAACCAGGGCGGCGCGCGTCCGGCGCGCGGCAGACGGTGGGCGGGCAGTTCCGCGCGTCGCTGTCGGCGCTGATGCTGACGCTGTCCCGCACCACGCCCCACTACGTGCGCTGCATCAAGCCCAACGACTCCAAGACGCCCTTCCTGTTCGACTCCGCGCGGGCCGTCAACCAGCTCAGGGCTTGCGGCGTGCTGGAGACGATACGGATATCTGCAGCCGGCTTCCCCTCCAGGTGGCTGTACCAGGACTTCTTCCATCGATACCGCCTCCTCTGTCGGCACAAGGACATCGACCGCGCCAACATCAAAGGGACGTGCTCCATAATTCTTTCCAAGCACCTCAAGGATCCGGACAAATACCAGTTCGGTGCCACGAAGATATTCTTCCGGGCGGGACAGGTCGCATACTTAGAGAAGGTCCGCGCGGACATACAGCGCGAGTACTGCGTGCGCGTGCAGAGCTGCGTGCGCACGTTCGTGGCCCGTCGGAAGTACCTCCGGGTCATGAGGACCGTGGTCGGCATCCAGGCCCGCTCCCGGGGCTACCTGGCCAGGCGCAAAGCGCAGGAGCTACGCCGCGCCCGCGCTGCCGTGACCCTCCAGCGGCACGTGCGCGGGTGGCTGGCGCGGCGCAACTACCGCCGGCTGCGGGGCCTGGCGCTGGCGCTGCAGGCGCGCGCCCGCGGGTACCTCGCCAGGAGACTGTACCGGGACCGCAGGAAACTAAGAGCCGTCGTCACGATACAGAGATACGCTCGCGGCTTCTTGGCTAGACAGAGGGTGAAGAAGATGAAGAGACAGATCATAGTCGCGCAGGCAGCGATCCGTCGTTTCCTAGCGCGTCGCCAGTACAAGCGGCTCCGCATCGAGGCCCGCAGCCTCGACCACGTGAAGACCCTCAACAAGGGCCTGGAGAACAAGATCATCAGCCTGCAGCAGCGGCTCGGCGGCGCCATGGAGAAGAACAAGCTGATCGAGCCGCTCACCGCGCAGGTCGCGGACCTCAAGGCAAAACTGGAACTACTCAAACTGGTTGAGATCGAAGTGAAAACGTTAAAAGTCGGCATCGACGACAAGGACAGCATTATAAACGCGCTGCGAGCGGAACTGCAGTCGGAAAGGGACGCCAATACGCGACTCGCCCAAGCGAAGAAAGGGATAGAGGATCAATATAACAAGGACAGACTGACTTGGGAAGAAGAGAGCGAGAAATTGGCCAACGAACTAAAGAGCGTCAAAGAAAACTACACGTTAGCTATTGAAGAACGGGATAGGCAACATGAGTTGGAGAAGAGTAAACTCAGTGCCGATTTAGAATCGGAGAGGCAGAGTCGTCAGAAGCTGCTGTCCTCGCAGTATGAACTCCAGGAACGTATTGACACGCTACAGAGGGCGCCGCCGGCCAAGGAACACAGGAGGTCGCTCTCCGACGCCAGCAGCAACTCGCAGCAGGAGACCGCAGTCGAGGATGACTACGGCTACGGGTCGGTGCGGTCCGTGGACACCAGCAGGCCCGCCCTGGAGGCCGTGAACTGGTCCGCGGGCCACAACAACGGGACCGTGCCGGTGGCGGACGCCGGGCTGGTGTTGCGCATGCAGAACAGGATCTCGGCGCTGCAGAGCGAGCTGTCCCGGACCACCAAGCGCGCCGCCGACCTGGAGGAGAGGCTGCTCAGTTAG
Protein
MSVQQPGRRASGARQTVGGQFRASLSALMLTLSRTTPHYVRCIKPNDSKTPFLFDSARAVNQLRACGVLETIRISAAGFPSRWLYQDFFHRYRLLCRHKDIDRANIKGTCSIILSKHLKDPDKYQFGATKIFFRAGQVAYLEKVRADIQREYCVRVQSCVRTFVARRKYLRVMRTVVGIQARSRGYLARRKAQELRRARAAVTLQRHVRGWLARRNYRRLRGLALALQARARGYLARRLYRDRRKLRAVVTIQRYARGFLARQRVKKMKRQIIVAQAAIRRFLARRQYKRLRIEARSLDHVKTLNKGLENKIISLQQRLGGAMEKNKLIEPLTAQVADLKAKLELLKLVEIEVKTLKVGIDDKDSIINALRAELQSERDANTRLAQAKKGIEDQYNKDRLTWEEESEKLANELKSVKENYTLAIEERDRQHELEKSKLSADLESERQSRQKLLSSQYELQERIDTLQRAPPAKEHRRSLSDASSNSQQETAVEDDYGYGSVRSVDTSRPALEAVNWSAGHNNGTVPVADAGLVLRMQNRISALQSELSRTTKRAADLEERLLS
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
H9JJS8
A0A2A4JEB0
A0A194PUG4
U4UQP3
N6TJX6
A0A0T6BA34
+ More
D6WQH7 A0A1Y1MHK0 A0A0J7L868 E0VAZ3 A0A026WN41 T1JHV8 A0A1B6FZ72 T1PMX1 A0A069DY44 A0A1I8MPE3 A0A1I8MPE1 A0A154P7I5 A0A2M4CPU8 A0A1Q3G1V7 A0A3B1IEH2 A0A2M4AAB6 A0A182G783 Q16W30 A0A1S4FM37 A0A182FGK3 A0A2M4A7I6 A0A2D0QVM5 A0A2D0QTA2 W5JMF7 G9KC72 A0A3L8DE04 A0A3B3QCH2 A0A1W4Z392 A0A1W4Z380 A0A1W4Z3M4 A0A1W4YUB2 F7EFH9 A0A1W4Z3L9 A0A1W4YUA6 A0A1W4Z387 A0A1L8GSX4 H3A8H4 A0A182QG83 A0A1L8H0F8 A0A182NJ12 K4JEU1 A0A0P6JX10 G1STJ8 G5C9D1 J9P789 U6DJ78 J9P0Z2 F1Q4E3 K7FSJ0 M3Y858 A0A091QXE5 A0A182YFJ3 H0WNA0 F7AXX3 A0A091CUQ2 F7AXY3 F7B2Y1 I3M966 A0A3Q0DX86 A0A2Y9IRH9 A0A2Y9IIW7 A0A2Y9IK38 A0A182RR43 S9WAX5 A0A093JEB9 G1LLE2 U3ICS9 G1LLD1 D2HN72 A0A2K6FYE5 A0A1V4JE98 A0A2K6FYD9 A0A182PPF8 A0A0D9RGH7 G3GRI6 A0A182K033 A0A384DBV2 Q59FF5 A0A2I0MDL4 A0A2K5PC40 F6ZG17 U3D823 A0A2K5PBT3 A0A2K5D2W8 A0A2K5PBV0 F7I698 U3B9M8
D6WQH7 A0A1Y1MHK0 A0A0J7L868 E0VAZ3 A0A026WN41 T1JHV8 A0A1B6FZ72 T1PMX1 A0A069DY44 A0A1I8MPE3 A0A1I8MPE1 A0A154P7I5 A0A2M4CPU8 A0A1Q3G1V7 A0A3B1IEH2 A0A2M4AAB6 A0A182G783 Q16W30 A0A1S4FM37 A0A182FGK3 A0A2M4A7I6 A0A2D0QVM5 A0A2D0QTA2 W5JMF7 G9KC72 A0A3L8DE04 A0A3B3QCH2 A0A1W4Z392 A0A1W4Z380 A0A1W4Z3M4 A0A1W4YUB2 F7EFH9 A0A1W4Z3L9 A0A1W4YUA6 A0A1W4Z387 A0A1L8GSX4 H3A8H4 A0A182QG83 A0A1L8H0F8 A0A182NJ12 K4JEU1 A0A0P6JX10 G1STJ8 G5C9D1 J9P789 U6DJ78 J9P0Z2 F1Q4E3 K7FSJ0 M3Y858 A0A091QXE5 A0A182YFJ3 H0WNA0 F7AXX3 A0A091CUQ2 F7AXY3 F7B2Y1 I3M966 A0A3Q0DX86 A0A2Y9IRH9 A0A2Y9IIW7 A0A2Y9IK38 A0A182RR43 S9WAX5 A0A093JEB9 G1LLE2 U3ICS9 G1LLD1 D2HN72 A0A2K6FYE5 A0A1V4JE98 A0A2K6FYD9 A0A182PPF8 A0A0D9RGH7 G3GRI6 A0A182K033 A0A384DBV2 Q59FF5 A0A2I0MDL4 A0A2K5PC40 F6ZG17 U3D823 A0A2K5PBT3 A0A2K5D2W8 A0A2K5PBV0 F7I698 U3B9M8
Pubmed
19121390
26354079
23537049
18362917
19820115
28004739
+ More
20566863 24508170 26334808 25315136 25329095 26483478 17510324 20920257 23761445 23236062 30249741 29240929 20431018 27762356 9215903 21993624 21993625 16341006 17381049 25244985 19892987 23149746 20010809 23749191 21804562 23371554 25243066
20566863 24508170 26334808 25315136 25329095 26483478 17510324 20920257 23761445 23236062 30249741 29240929 20431018 27762356 9215903 21993624 21993625 16341006 17381049 25244985 19892987 23149746 20010809 23749191 21804562 23371554 25243066
EMBL
BABH01017761
BABH01017762
NWSH01001891
PCG69780.1
KQ459593
KPI96618.1
+ More
KB632326 ERL92471.1 APGK01035149 APGK01035150 APGK01035151 KB740923 ENN78188.1 LJIG01002798 KRT84142.1 KQ971354 EFA06975.2 GEZM01031022 JAV85121.1 LBMM01000310 KMR01422.1 DS235019 EEB10549.1 KK107148 EZA57445.1 JH431845 GECZ01014290 JAS55479.1 KA650131 AFP64760.1 GBGD01000079 JAC88810.1 KQ434822 KZC07080.1 GGFL01003159 MBW67337.1 GFDL01001250 JAV33795.1 GGFK01004384 MBW37705.1 JXUM01047023 JXUM01047024 JXUM01047025 JXUM01047026 JXUM01047027 JXUM01047028 CH477576 EAT38792.1 GGFK01003446 MBW36767.1 ADMH02001146 ETN63934.1 JP013899 AES02497.1 QOIP01000009 RLU18383.1 AAMC01056619 AAMC01056620 AAMC01056621 CM004471 OCT86947.1 AFYH01203852 AFYH01203853 AXCN02000461 CM004470 OCT89580.1 JX494698 AFU81219.2 GEBF01002293 JAO01340.1 AAGW02024800 AAGW02024801 AAGW02024802 AAGW02024803 AAGW02024804 AAGW02024805 AAGW02024806 AAGW02024807 AAGW02024808 AAGW02024809 JH174048 EHB18142.1 AAEX03016140 HAAF01010820 CCP82644.1 AGCU01054991 AGCU01054992 AGCU01054993 AGCU01054994 AGCU01054995 AEYP01039009 AEYP01039010 AEYP01039011 AEYP01039012 AEYP01039013 AEYP01039014 AEYP01039015 AEYP01039016 KK709155 KFQ32240.1 AAQR03024867 AAQR03024868 AAQR03024869 AAQR03024870 AAQR03024871 AAQR03024872 AAQR03024873 AAQR03024874 AAQR03024875 AAQR03024876 KN124379 KFO21415.1 AGTP01002747 AGTP01002748 AGTP01002749 KB019205 EPY73094.1 KK575648 KFW10242.1 ACTA01044279 ACTA01052279 ACTA01060279 ACTA01068279 ADON01084796 ADON01084797 ADON01084798 GL193076 EFB21521.1 LSYS01007908 OPJ70593.1 AQIB01116024 AQIB01116025 AQIB01116026 AQIB01116027 AQIB01116028 JH000003 EGV99862.1 AB209487 AB209505 BAD92742.1 AKCR02000019 PKK27768.1 GAMT01002420 GAMT01002419 JAB09441.1 GAMT01002418 GAMS01006862 JAB09443.1 JAB16274.1
KB632326 ERL92471.1 APGK01035149 APGK01035150 APGK01035151 KB740923 ENN78188.1 LJIG01002798 KRT84142.1 KQ971354 EFA06975.2 GEZM01031022 JAV85121.1 LBMM01000310 KMR01422.1 DS235019 EEB10549.1 KK107148 EZA57445.1 JH431845 GECZ01014290 JAS55479.1 KA650131 AFP64760.1 GBGD01000079 JAC88810.1 KQ434822 KZC07080.1 GGFL01003159 MBW67337.1 GFDL01001250 JAV33795.1 GGFK01004384 MBW37705.1 JXUM01047023 JXUM01047024 JXUM01047025 JXUM01047026 JXUM01047027 JXUM01047028 CH477576 EAT38792.1 GGFK01003446 MBW36767.1 ADMH02001146 ETN63934.1 JP013899 AES02497.1 QOIP01000009 RLU18383.1 AAMC01056619 AAMC01056620 AAMC01056621 CM004471 OCT86947.1 AFYH01203852 AFYH01203853 AXCN02000461 CM004470 OCT89580.1 JX494698 AFU81219.2 GEBF01002293 JAO01340.1 AAGW02024800 AAGW02024801 AAGW02024802 AAGW02024803 AAGW02024804 AAGW02024805 AAGW02024806 AAGW02024807 AAGW02024808 AAGW02024809 JH174048 EHB18142.1 AAEX03016140 HAAF01010820 CCP82644.1 AGCU01054991 AGCU01054992 AGCU01054993 AGCU01054994 AGCU01054995 AEYP01039009 AEYP01039010 AEYP01039011 AEYP01039012 AEYP01039013 AEYP01039014 AEYP01039015 AEYP01039016 KK709155 KFQ32240.1 AAQR03024867 AAQR03024868 AAQR03024869 AAQR03024870 AAQR03024871 AAQR03024872 AAQR03024873 AAQR03024874 AAQR03024875 AAQR03024876 KN124379 KFO21415.1 AGTP01002747 AGTP01002748 AGTP01002749 KB019205 EPY73094.1 KK575648 KFW10242.1 ACTA01044279 ACTA01052279 ACTA01060279 ACTA01068279 ADON01084796 ADON01084797 ADON01084798 GL193076 EFB21521.1 LSYS01007908 OPJ70593.1 AQIB01116024 AQIB01116025 AQIB01116026 AQIB01116027 AQIB01116028 JH000003 EGV99862.1 AB209487 AB209505 BAD92742.1 AKCR02000019 PKK27768.1 GAMT01002420 GAMT01002419 JAB09441.1 GAMT01002418 GAMS01006862 JAB09443.1 JAB16274.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000030742
UP000019118
UP000007266
+ More
UP000036403 UP000009046 UP000053097 UP000095301 UP000076502 UP000018467 UP000069940 UP000008820 UP000069272 UP000221080 UP000000673 UP000279307 UP000261540 UP000192224 UP000008143 UP000186698 UP000008672 UP000075886 UP000075884 UP000001811 UP000006813 UP000002254 UP000007267 UP000000715 UP000076408 UP000005225 UP000002281 UP000028990 UP000005215 UP000189704 UP000248482 UP000075900 UP000008912 UP000016666 UP000233160 UP000190648 UP000075885 UP000029965 UP000001075 UP000075881 UP000261680 UP000053872 UP000233040 UP000008225 UP000233020
UP000036403 UP000009046 UP000053097 UP000095301 UP000076502 UP000018467 UP000069940 UP000008820 UP000069272 UP000221080 UP000000673 UP000279307 UP000261540 UP000192224 UP000008143 UP000186698 UP000008672 UP000075886 UP000075884 UP000001811 UP000006813 UP000002254 UP000007267 UP000000715 UP000076408 UP000005225 UP000002281 UP000028990 UP000005215 UP000189704 UP000248482 UP000075900 UP000008912 UP000016666 UP000233160 UP000190648 UP000075885 UP000029965 UP000001075 UP000075881 UP000261680 UP000053872 UP000233040 UP000008225 UP000233020
Interpro
Gene 3D
ProteinModelPortal
H9JJS8
A0A2A4JEB0
A0A194PUG4
U4UQP3
N6TJX6
A0A0T6BA34
+ More
D6WQH7 A0A1Y1MHK0 A0A0J7L868 E0VAZ3 A0A026WN41 T1JHV8 A0A1B6FZ72 T1PMX1 A0A069DY44 A0A1I8MPE3 A0A1I8MPE1 A0A154P7I5 A0A2M4CPU8 A0A1Q3G1V7 A0A3B1IEH2 A0A2M4AAB6 A0A182G783 Q16W30 A0A1S4FM37 A0A182FGK3 A0A2M4A7I6 A0A2D0QVM5 A0A2D0QTA2 W5JMF7 G9KC72 A0A3L8DE04 A0A3B3QCH2 A0A1W4Z392 A0A1W4Z380 A0A1W4Z3M4 A0A1W4YUB2 F7EFH9 A0A1W4Z3L9 A0A1W4YUA6 A0A1W4Z387 A0A1L8GSX4 H3A8H4 A0A182QG83 A0A1L8H0F8 A0A182NJ12 K4JEU1 A0A0P6JX10 G1STJ8 G5C9D1 J9P789 U6DJ78 J9P0Z2 F1Q4E3 K7FSJ0 M3Y858 A0A091QXE5 A0A182YFJ3 H0WNA0 F7AXX3 A0A091CUQ2 F7AXY3 F7B2Y1 I3M966 A0A3Q0DX86 A0A2Y9IRH9 A0A2Y9IIW7 A0A2Y9IK38 A0A182RR43 S9WAX5 A0A093JEB9 G1LLE2 U3ICS9 G1LLD1 D2HN72 A0A2K6FYE5 A0A1V4JE98 A0A2K6FYD9 A0A182PPF8 A0A0D9RGH7 G3GRI6 A0A182K033 A0A384DBV2 Q59FF5 A0A2I0MDL4 A0A2K5PC40 F6ZG17 U3D823 A0A2K5PBT3 A0A2K5D2W8 A0A2K5PBV0 F7I698 U3B9M8
D6WQH7 A0A1Y1MHK0 A0A0J7L868 E0VAZ3 A0A026WN41 T1JHV8 A0A1B6FZ72 T1PMX1 A0A069DY44 A0A1I8MPE3 A0A1I8MPE1 A0A154P7I5 A0A2M4CPU8 A0A1Q3G1V7 A0A3B1IEH2 A0A2M4AAB6 A0A182G783 Q16W30 A0A1S4FM37 A0A182FGK3 A0A2M4A7I6 A0A2D0QVM5 A0A2D0QTA2 W5JMF7 G9KC72 A0A3L8DE04 A0A3B3QCH2 A0A1W4Z392 A0A1W4Z380 A0A1W4Z3M4 A0A1W4YUB2 F7EFH9 A0A1W4Z3L9 A0A1W4YUA6 A0A1W4Z387 A0A1L8GSX4 H3A8H4 A0A182QG83 A0A1L8H0F8 A0A182NJ12 K4JEU1 A0A0P6JX10 G1STJ8 G5C9D1 J9P789 U6DJ78 J9P0Z2 F1Q4E3 K7FSJ0 M3Y858 A0A091QXE5 A0A182YFJ3 H0WNA0 F7AXX3 A0A091CUQ2 F7AXY3 F7B2Y1 I3M966 A0A3Q0DX86 A0A2Y9IRH9 A0A2Y9IIW7 A0A2Y9IK38 A0A182RR43 S9WAX5 A0A093JEB9 G1LLE2 U3ICS9 G1LLD1 D2HN72 A0A2K6FYE5 A0A1V4JE98 A0A2K6FYD9 A0A182PPF8 A0A0D9RGH7 G3GRI6 A0A182K033 A0A384DBV2 Q59FF5 A0A2I0MDL4 A0A2K5PC40 F6ZG17 U3D823 A0A2K5PBT3 A0A2K5D2W8 A0A2K5PBV0 F7I698 U3B9M8
PDB
2DFS
E-value=9.10341e-77,
Score=732
Ontologies
GO
GO:0005524
GO:0016459
GO:0051015
GO:0003774
GO:0003779
GO:0016461
GO:0005777
GO:0005884
GO:0032402
GO:0042759
GO:0097718
GO:0072659
GO:0005509
GO:0055037
GO:0042438
GO:0030318
GO:0001750
GO:0006892
GO:0042470
GO:0032433
GO:0005770
GO:0031585
GO:0007601
GO:0017137
GO:0098978
GO:0042802
GO:0016020
GO:0032593
GO:0031987
GO:0042476
GO:0032869
GO:0030050
GO:0035371
GO:0006887
GO:0099566
GO:0001726
GO:0007268
GO:0043025
GO:0005829
GO:0098794
GO:0005790
GO:0032252
GO:0005764
GO:0005516
GO:0000146
GO:0030073
GO:0048820
GO:0042641
GO:0099089
GO:0005794
GO:0050808
GO:0005769
GO:0042552
GO:0005882
GO:0005515
GO:0005737
GO:0006396
GO:0000166
GO:0004887
GO:0003707
GO:0016021
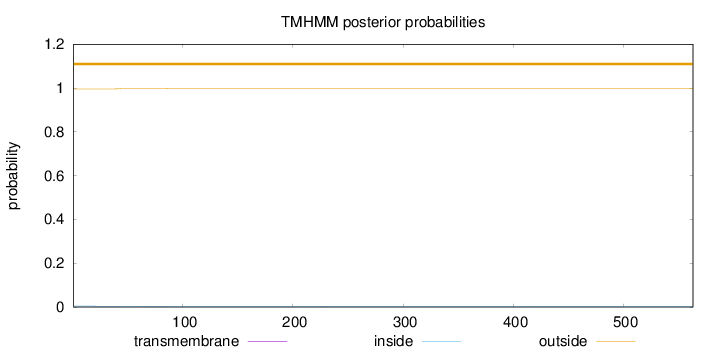
Topology
Length:
563
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0487200000000003
Exp number, first 60 AAs:
0.02149
Total prob of N-in:
0.00452
outside
1 - 563
Population Genetic Test Statistics
Pi
275.110078
Theta
156.329052
Tajima's D
2.116394
CLR
0.05262
CSRT
0.901654917254137
Interpretation
Uncertain
