Pre Gene Modal
BGIBMGA009779
Annotation
PREDICTED:_unconventional_myosin-Va_[Bombyx_mori]
Full name
Unconventional myosin-Vb
+ More
Unconventional myosin-Va
Unconventional myosin-Va
Alternative Name
Dilute myosin heavy chain, non-muscle
Myosin heavy chain p190
Myosin-V
Myosin heavy chain p190
Myosin-V
Location in the cell
Cytoplasmic Reliability : 1.941
Sequence
CDS
ATGACAACGAAGGAATTGTACACGAAGGGTGCCAGGATATGGGTGGAGCACCCGGAGAAGGTGTGGGAGAGCGCAACAGTAACATCCGACTATCGATCCGGCGTTCTCATCGTGAAACTAGACCAGACAGATGAAATAAGACAAATAACGATCAAAGATGAGAAACACATGCCCCCACTCCGGAACCCGTCGCTCCTCATCGGCCAAAATGATCTCACGTCACTATCGTACCTCCACGAGCCGGCGGTGCTGCATAATCTTAAAGTCAGATTCTGCGATCGTAACGCTATATACACCTACTGCGGGATCGTCCTCGTGGCCATCAATCCTTATTATGATCTTCCGATTTACGGCGATGAAACCATCATGGCGTATCGGGGCCAATCGATGGGCGATCTGGACCCTCACATATTCGCCGTCTCCGAAGAGGCCTACACCAAACTGGAGCGAGAAAGGAGTGATCAGAGTATCATAGTCAGCGGCGAGTCCGGTGCCGGGAAGACGGTGTCGGCAAAGTACGCCATGCGATACTTCGCAGCTGTCGGCGGCAACGTTAGCGAAACCCAGATCGAGAGGAAAGTGCTGGCTTCAAGTCCCATCATGGAGGCGATAGGCAATGCGAAGACAACCCGTAACGACAACTCATCGCGCTTCGGCAAGTTCATAGAGATACACTTCGACAGCCAGTACCGGATATCCGGGGCCAGCATGCGCACGTACCTGCTCGAGAAGTCGAGGGTGGTGTACCAGAGTCCCGGCGAGAGGAACTACCATATATTCTATCAGCTGTGTGCCGCTTCTGCTTTGATGCCTCATCTGAAGTTGGAACATCAAGACACATTACACTATTTGAATCAAGGCGGCAGTCCGAATATCGACGGAGTGGATGATCTAAAAGCTTTCAACGAAACTAGAAGCGCTTTAACTACTTTAGGCGTTACGGATTCGGAGCAGAGGGACATGTTCACCATCCTGGCCGCGATACTGCATCTCGGCAACGTCGCGCTGCTGTCCGAGGACCCCGACGCGGCGCCCGGAGCAGACTGCGAGGACGGAGCCTACATCAGCCCGACAGACGAGCACCTGACGCACGCGTGTTCGCTGCTGGGAGTGTCGCGCTTGGAGCTGTCGCGGTGGCTGAGTCATCGCCGCATCGCCTCCGCCCACGAGGTCATCGTCGCCAGGCTGGACGTGCAGCGCGCGGCGTGCGCCCGGGACGCGCTCGCCAAGAAGCTGTACGGGGAGCTGTTCGCGTGGCTCGTCACCGCAGTCAACCGCGCGCTCGACACCGGACACGCCAAGAAACACTTCATCGGCGTCCTCGACATCTACGGCTTCGAGACCTTCGAAATCAACTCGTTCGAACAGTTCTGCATCAACTACGCGAACGAGAAGCTCCAGCAGCAGTTCAACTCGCACGTGTTCAAACTCGAGCAGGACGAATATATAAAGGAAGAGATCTCGTGGGAGATGATTGACTTCTACGACAACCAGCCGTGCATAGACCTGATCGAGGACCGTCTGGGTGTCCTGGCGCTGCTCGACGAGGAGTGCCGCGTGCCGCAGGGCTCCGACACCGGCTTCGTGGCCAAGCTGCACCAGAAGTGCGCCGCCTACCCGCACTTCATGAAGCCGAGGTTCGGGAACCAGGCCTTCATAATAAAGCATTTCGCTGACAACGTGGAGTACCAGTCCGGCGGTTTCCTTGAGAAGAATCGTGACACAGTGTCGGAGGAACAGCTGGAGTGCATCAGGACAGCCACCAGCTGCCGCCTCATACACGTCATGCTGGAGTCGGAGCGCGCGGTAGACCGTCAGACCGCGACCCTGCCGCCGCCCTCCCGGAGGAGGACCACGCCCTCCGTGCCGATAACGAGTCTAACTAAAATCCAACTCAAAAATAGAAAATAA
Protein
MTTKELYTKGARIWVEHPEKVWESATVTSDYRSGVLIVKLDQTDEIRQITIKDEKHMPPLRNPSLLIGQNDLTSLSYLHEPAVLHNLKVRFCDRNAIYTYCGIVLVAINPYYDLPIYGDETIMAYRGQSMGDLDPHIFAVSEEAYTKLERERSDQSIIVSGESGAGKTVSAKYAMRYFAAVGGNVSETQIERKVLASSPIMEAIGNAKTTRNDNSSRFGKFIEIHFDSQYRISGASMRTYLLEKSRVVYQSPGERNYHIFYQLCAASALMPHLKLEHQDTLHYLNQGGSPNIDGVDDLKAFNETRSALTTLGVTDSEQRDMFTILAAILHLGNVALLSEDPDAAPGADCEDGAYISPTDEHLTHACSLLGVSRLELSRWLSHRRIASAHEVIVARLDVQRAACARDALAKKLYGELFAWLVTAVNRALDTGHAKKHFIGVLDIYGFETFEINSFEQFCINYANEKLQQQFNSHVFKLEQDEYIKEEISWEMIDFYDNQPCIDLIEDRLGVLALLDEECRVPQGSDTGFVAKLHQKCAAYPHFMKPRFGNQAFIIKHFADNVEYQSGGFLEKNRDTVSEEQLECIRTATSCRLIHVMLESERAVDRQTATLPPPSRRRTTPSVPITSLTKIQLKNRK
Summary
Description
May be involved in vesicular trafficking via its association with the CART complex. The CART complex is necessary for efficient transferrin receptor recycling but not for EGFR degradation. Required in a complex with RAB11A and RAB11FIP2 for the transport of NPC1L1 to the plasma membrane. Together with RAB11A participates in CFTR trafficking to the plasma membrane and TF (transferrin) recycling in nonpolarized cells. Together with RAB11A and RAB8A participates in epithelial cell polarization. Together with RAB25 regulates transcytosis.
Processive actin-based motor that can move in large steps approximating the 36-nm pseudo-repeat of the actin filament. Involved in melanosome transport. Also mediates the transport of vesicles to the plasma membrane. May also be required for some polarization process involved in dendrite formation (By similarity).
Processive actin-based motor that can move in large steps approximating the 36-nm pseudo-repeat of the actin filament. Involved in melanosome transport. Also mediates the transport of vesicles to the plasma membrane. May also be required for some polarization process involved in dendrite formation (By similarity).
Subunit
Component of the CART complex, at least composed of ACTN4, HGS/HRS, MYO5B and TRIM3. Interacts with RAB11FIP2 (By similarity). Interacts with RAB11A and RAB8A (By similarity). Found in a complex with CFTR and RAB11A (By similarity). Interacts with NPC1L1 (By similarity). Interacts with LIMA1 (PubMed:29880681).
May be a homodimer, which associates with multiple calmodulin or myosin light chains.
May be a homodimer, which associates with multiple calmodulin or myosin light chains.
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Keywords
Actin-binding
Alternative splicing
ATP-binding
Calmodulin-binding
Coiled coil
Complete proteome
Cytoplasm
Motor protein
Myosin
Nucleotide-binding
Phosphoprotein
Protein transport
Reference proteome
Repeat
Transport
3D-structure
Direct protein sequencing
Golgi apparatus
Membrane
Feature
chain Unconventional myosin-Vb
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9JJS8
A0A194PUG4
A0A2H1V2L1
D6WQH7
V5G7A5
K7IV32
+ More
A0A088AGS8 A0A1B6FZ72 A0A195CUA2 V9IC23 A0A3L8DE04 A0A158NQ81 A0A1Y1MHK0 A0A154P7I5 E2AL94 A0A232FB52 A0A0N0U3P1 A0A0J7L868 A0A195FNW2 A0A1B6I9C2 A0A2A3E8F5 A0A151WYN4 A0A0L7R6N9 A0A0L7R6Y3 A0A195DEK4 E2A5K9 A0A026WN41 E2B2T6 A0A0P4W5R4 A0A0P4WGV2 F6K356 U4UQP3 N6TJX6 A0A0F7VJF0 A0A0C9QSS3 A0A0C9QTC4 A0A182QG83 E0VAZ3 A0A195BJ13 A0A182MMU4 A0A0A1WZ66 A0A0A1X3X1 A0A0K8W7J1 A0A0K8U9G9 A0A182LKT9 A0A0K2VG83 W8B8Y3 W8B4M8 A0A0P5P9E7 A0A0P4Y350 A0A0P6HK55 A0A0P6FW99 A0A0P5LPI2 A0A0N8CXS1 A0A0P5IF88 A0A0P5VR14 A0A0P5RKE3 A0A0P5RPT9 A0A0N8C7R8 A0A3B0J347 A0A0N8CE72 E9H736 A0A3B0J2M3 A0A0K8SQI6 A0A0A9ZGU3 A0A0P6CNX1 A0A182JIC1 A0A0A1X772 A0A0P6EJY1 A0A0Q9XLC5 B4KQ96 A0A0K8WJ51 A0A1L8I2T7 A0A1Q3G1V7 B4H8K7 B4J9Z7 P21271 P21271-3 Q28YI2 N6W4L9 A0A0T6BA34 A0A0Q9XHH8 F6WHW2 A0A182G783 A0A182RR43 Q02440 A0A1D5PRP3 A0A0P6BUJ9 A0A182V3H7 A0A182I464 G1PV90 A0A182WC09 A0A1B6DXB9 A0A2M4CPU8 A0A3Q0CRB3 G5E8G6 Q7Q5E6 G3X9Y9 A0A0L0CAC5
A0A088AGS8 A0A1B6FZ72 A0A195CUA2 V9IC23 A0A3L8DE04 A0A158NQ81 A0A1Y1MHK0 A0A154P7I5 E2AL94 A0A232FB52 A0A0N0U3P1 A0A0J7L868 A0A195FNW2 A0A1B6I9C2 A0A2A3E8F5 A0A151WYN4 A0A0L7R6N9 A0A0L7R6Y3 A0A195DEK4 E2A5K9 A0A026WN41 E2B2T6 A0A0P4W5R4 A0A0P4WGV2 F6K356 U4UQP3 N6TJX6 A0A0F7VJF0 A0A0C9QSS3 A0A0C9QTC4 A0A182QG83 E0VAZ3 A0A195BJ13 A0A182MMU4 A0A0A1WZ66 A0A0A1X3X1 A0A0K8W7J1 A0A0K8U9G9 A0A182LKT9 A0A0K2VG83 W8B8Y3 W8B4M8 A0A0P5P9E7 A0A0P4Y350 A0A0P6HK55 A0A0P6FW99 A0A0P5LPI2 A0A0N8CXS1 A0A0P5IF88 A0A0P5VR14 A0A0P5RKE3 A0A0P5RPT9 A0A0N8C7R8 A0A3B0J347 A0A0N8CE72 E9H736 A0A3B0J2M3 A0A0K8SQI6 A0A0A9ZGU3 A0A0P6CNX1 A0A182JIC1 A0A0A1X772 A0A0P6EJY1 A0A0Q9XLC5 B4KQ96 A0A0K8WJ51 A0A1L8I2T7 A0A1Q3G1V7 B4H8K7 B4J9Z7 P21271 P21271-3 Q28YI2 N6W4L9 A0A0T6BA34 A0A0Q9XHH8 F6WHW2 A0A182G783 A0A182RR43 Q02440 A0A1D5PRP3 A0A0P6BUJ9 A0A182V3H7 A0A182I464 G1PV90 A0A182WC09 A0A1B6DXB9 A0A2M4CPU8 A0A3Q0CRB3 G5E8G6 Q7Q5E6 G3X9Y9 A0A0L0CAC5
Pubmed
19121390
26354079
18362917
19820115
20075255
30249741
+ More
21347285 28004739 20798317 28648823 24508170 22846366 23537049 20566863 25830018 20966253 24495485 21292972 25401762 17994087 27762356 15489334 15368895 2236059 21183079 29880681 15632085 20431018 26483478 1383040 1469047 21993624 26319212 12040188 19468303 12364791 26108605
21347285 28004739 20798317 28648823 24508170 22846366 23537049 20566863 25830018 20966253 24495485 21292972 25401762 17994087 27762356 15489334 15368895 2236059 21183079 29880681 15632085 20431018 26483478 1383040 1469047 21993624 26319212 12040188 19468303 12364791 26108605
EMBL
BABH01017761
BABH01017762
KQ459593
KPI96618.1
ODYU01000382
SOQ35071.1
+ More
KQ971354 EFA06975.2 GALX01002476 JAB65990.1 AAZX01001194 GECZ01014290 JAS55479.1 KQ977279 KYN04261.1 JR039293 JR039294 AEY58619.1 AEY58620.1 QOIP01000009 RLU18383.1 ADTU01022924 ADTU01022925 GEZM01031022 JAV85121.1 KQ434822 KZC07080.1 GL440553 EFN65790.1 NNAY01000532 OXU27842.1 KQ435896 KOX69324.1 LBMM01000310 KMR01422.1 KQ981382 KYN42143.1 GECU01024162 JAS83544.1 KZ288326 PBC27980.1 KQ982652 KYQ52811.1 KQ414646 KOC66513.1 KOC66516.1 KQ980934 KYN11323.1 GL436993 EFN71292.1 KK107148 EZA57445.1 GL445235 EFN90001.1 GDRN01089957 JAI60556.1 GDRN01089955 JAI60557.1 HM212671 ADM15669.1 KB632326 ERL92471.1 APGK01035149 APGK01035150 APGK01035151 KB740923 ENN78188.1 LN830727 CFW94236.1 GBYB01003692 JAG73459.1 GBYB01006959 JAG76726.1 AXCN02000461 DS235019 EEB10549.1 KQ976464 KYM84426.1 AXCM01004568 GBXI01010584 JAD03708.1 GBXI01008248 JAD06044.1 GDHF01005267 JAI47047.1 GDHF01029304 JAI23010.1 HACA01031851 CDW49212.1 GAMC01013037 JAB93518.1 GAMC01013038 JAB93517.1 GDIQ01141357 JAL10369.1 GDIP01235456 JAI87945.1 GDIQ01018624 JAN76113.1 GDIQ01041893 JAN52844.1 GDIQ01170346 GDIQ01136811 GDIQ01030299 LRGB01003163 JAK81379.1 KZS03930.1 GDIP01088479 JAM15236.1 GDIQ01214230 JAK37495.1 GDIP01096726 JAM06989.1 GDIQ01108817 JAL42909.1 GDIQ01106568 JAL45158.1 GDIQ01106515 JAL45211.1 OUUW01000001 SPP75705.1 GDIP01140471 JAL63243.1 GL732599 EFX72449.1 SPP75704.1 GBRD01010430 GBRD01007165 JAG55394.1 GBHO01000214 GBHO01000213 JAG43390.1 JAG43391.1 GDIQ01090022 JAN04715.1 GBXI01007769 JAD06523.1 GDIQ01061683 JAN33054.1 CH933808 KRG04531.1 EDW09224.1 GDHF01001133 JAI51181.1 CM004466 OCU02687.1 GFDL01001250 JAV33795.1 CH479223 EDW35042.1 CH916367 EDW02584.1 BC011494 BC046444 BC118525 AK173102 M55253 CM000071 EAL25983.2 ENO01762.2 LJIG01002798 KRT84142.1 KRG04530.1 AAMC01059172 AAMC01059173 AAMC01059174 AAMC01059175 AAMC01059176 AAMC01059177 AAMC01059178 AAMC01059179 JXUM01047023 JXUM01047024 JXUM01047025 JXUM01047026 JXUM01047027 JXUM01047028 X67251 Z11718 GDIP01017260 JAM86455.1 APCN01002280 AAPE02009051 AAPE02009052 GEDC01028278 GEDC01006987 JAS09020.1 JAS30311.1 GGFL01003159 MBW67337.1 AC147992 AC148001 AC148002 CH466528 EDL09524.1 AAAB01008960 EAA11777.4 EDL09522.1 JRES01000780 KNC28384.1
KQ971354 EFA06975.2 GALX01002476 JAB65990.1 AAZX01001194 GECZ01014290 JAS55479.1 KQ977279 KYN04261.1 JR039293 JR039294 AEY58619.1 AEY58620.1 QOIP01000009 RLU18383.1 ADTU01022924 ADTU01022925 GEZM01031022 JAV85121.1 KQ434822 KZC07080.1 GL440553 EFN65790.1 NNAY01000532 OXU27842.1 KQ435896 KOX69324.1 LBMM01000310 KMR01422.1 KQ981382 KYN42143.1 GECU01024162 JAS83544.1 KZ288326 PBC27980.1 KQ982652 KYQ52811.1 KQ414646 KOC66513.1 KOC66516.1 KQ980934 KYN11323.1 GL436993 EFN71292.1 KK107148 EZA57445.1 GL445235 EFN90001.1 GDRN01089957 JAI60556.1 GDRN01089955 JAI60557.1 HM212671 ADM15669.1 KB632326 ERL92471.1 APGK01035149 APGK01035150 APGK01035151 KB740923 ENN78188.1 LN830727 CFW94236.1 GBYB01003692 JAG73459.1 GBYB01006959 JAG76726.1 AXCN02000461 DS235019 EEB10549.1 KQ976464 KYM84426.1 AXCM01004568 GBXI01010584 JAD03708.1 GBXI01008248 JAD06044.1 GDHF01005267 JAI47047.1 GDHF01029304 JAI23010.1 HACA01031851 CDW49212.1 GAMC01013037 JAB93518.1 GAMC01013038 JAB93517.1 GDIQ01141357 JAL10369.1 GDIP01235456 JAI87945.1 GDIQ01018624 JAN76113.1 GDIQ01041893 JAN52844.1 GDIQ01170346 GDIQ01136811 GDIQ01030299 LRGB01003163 JAK81379.1 KZS03930.1 GDIP01088479 JAM15236.1 GDIQ01214230 JAK37495.1 GDIP01096726 JAM06989.1 GDIQ01108817 JAL42909.1 GDIQ01106568 JAL45158.1 GDIQ01106515 JAL45211.1 OUUW01000001 SPP75705.1 GDIP01140471 JAL63243.1 GL732599 EFX72449.1 SPP75704.1 GBRD01010430 GBRD01007165 JAG55394.1 GBHO01000214 GBHO01000213 JAG43390.1 JAG43391.1 GDIQ01090022 JAN04715.1 GBXI01007769 JAD06523.1 GDIQ01061683 JAN33054.1 CH933808 KRG04531.1 EDW09224.1 GDHF01001133 JAI51181.1 CM004466 OCU02687.1 GFDL01001250 JAV33795.1 CH479223 EDW35042.1 CH916367 EDW02584.1 BC011494 BC046444 BC118525 AK173102 M55253 CM000071 EAL25983.2 ENO01762.2 LJIG01002798 KRT84142.1 KRG04530.1 AAMC01059172 AAMC01059173 AAMC01059174 AAMC01059175 AAMC01059176 AAMC01059177 AAMC01059178 AAMC01059179 JXUM01047023 JXUM01047024 JXUM01047025 JXUM01047026 JXUM01047027 JXUM01047028 X67251 Z11718 GDIP01017260 JAM86455.1 APCN01002280 AAPE02009051 AAPE02009052 GEDC01028278 GEDC01006987 JAS09020.1 JAS30311.1 GGFL01003159 MBW67337.1 AC147992 AC148001 AC148002 CH466528 EDL09524.1 AAAB01008960 EAA11777.4 EDL09522.1 JRES01000780 KNC28384.1
Proteomes
UP000005204
UP000053268
UP000007266
UP000002358
UP000005203
UP000078542
+ More
UP000279307 UP000005205 UP000076502 UP000000311 UP000215335 UP000053105 UP000036403 UP000078541 UP000242457 UP000075809 UP000053825 UP000078492 UP000053097 UP000008237 UP000030742 UP000019118 UP000075886 UP000009046 UP000078540 UP000075883 UP000075882 UP000076858 UP000268350 UP000000305 UP000075880 UP000009192 UP000186698 UP000008744 UP000001070 UP000000589 UP000001819 UP000008143 UP000069940 UP000075900 UP000000539 UP000075903 UP000075840 UP000001074 UP000075920 UP000189706 UP000007062 UP000037069
UP000279307 UP000005205 UP000076502 UP000000311 UP000215335 UP000053105 UP000036403 UP000078541 UP000242457 UP000075809 UP000053825 UP000078492 UP000053097 UP000008237 UP000030742 UP000019118 UP000075886 UP000009046 UP000078540 UP000075883 UP000075882 UP000076858 UP000268350 UP000000305 UP000075880 UP000009192 UP000186698 UP000008744 UP000001070 UP000000589 UP000001819 UP000008143 UP000069940 UP000075900 UP000000539 UP000075903 UP000075840 UP000001074 UP000075920 UP000189706 UP000007062 UP000037069
Interpro
Gene 3D
ProteinModelPortal
H9JJS8
A0A194PUG4
A0A2H1V2L1
D6WQH7
V5G7A5
K7IV32
+ More
A0A088AGS8 A0A1B6FZ72 A0A195CUA2 V9IC23 A0A3L8DE04 A0A158NQ81 A0A1Y1MHK0 A0A154P7I5 E2AL94 A0A232FB52 A0A0N0U3P1 A0A0J7L868 A0A195FNW2 A0A1B6I9C2 A0A2A3E8F5 A0A151WYN4 A0A0L7R6N9 A0A0L7R6Y3 A0A195DEK4 E2A5K9 A0A026WN41 E2B2T6 A0A0P4W5R4 A0A0P4WGV2 F6K356 U4UQP3 N6TJX6 A0A0F7VJF0 A0A0C9QSS3 A0A0C9QTC4 A0A182QG83 E0VAZ3 A0A195BJ13 A0A182MMU4 A0A0A1WZ66 A0A0A1X3X1 A0A0K8W7J1 A0A0K8U9G9 A0A182LKT9 A0A0K2VG83 W8B8Y3 W8B4M8 A0A0P5P9E7 A0A0P4Y350 A0A0P6HK55 A0A0P6FW99 A0A0P5LPI2 A0A0N8CXS1 A0A0P5IF88 A0A0P5VR14 A0A0P5RKE3 A0A0P5RPT9 A0A0N8C7R8 A0A3B0J347 A0A0N8CE72 E9H736 A0A3B0J2M3 A0A0K8SQI6 A0A0A9ZGU3 A0A0P6CNX1 A0A182JIC1 A0A0A1X772 A0A0P6EJY1 A0A0Q9XLC5 B4KQ96 A0A0K8WJ51 A0A1L8I2T7 A0A1Q3G1V7 B4H8K7 B4J9Z7 P21271 P21271-3 Q28YI2 N6W4L9 A0A0T6BA34 A0A0Q9XHH8 F6WHW2 A0A182G783 A0A182RR43 Q02440 A0A1D5PRP3 A0A0P6BUJ9 A0A182V3H7 A0A182I464 G1PV90 A0A182WC09 A0A1B6DXB9 A0A2M4CPU8 A0A3Q0CRB3 G5E8G6 Q7Q5E6 G3X9Y9 A0A0L0CAC5
A0A088AGS8 A0A1B6FZ72 A0A195CUA2 V9IC23 A0A3L8DE04 A0A158NQ81 A0A1Y1MHK0 A0A154P7I5 E2AL94 A0A232FB52 A0A0N0U3P1 A0A0J7L868 A0A195FNW2 A0A1B6I9C2 A0A2A3E8F5 A0A151WYN4 A0A0L7R6N9 A0A0L7R6Y3 A0A195DEK4 E2A5K9 A0A026WN41 E2B2T6 A0A0P4W5R4 A0A0P4WGV2 F6K356 U4UQP3 N6TJX6 A0A0F7VJF0 A0A0C9QSS3 A0A0C9QTC4 A0A182QG83 E0VAZ3 A0A195BJ13 A0A182MMU4 A0A0A1WZ66 A0A0A1X3X1 A0A0K8W7J1 A0A0K8U9G9 A0A182LKT9 A0A0K2VG83 W8B8Y3 W8B4M8 A0A0P5P9E7 A0A0P4Y350 A0A0P6HK55 A0A0P6FW99 A0A0P5LPI2 A0A0N8CXS1 A0A0P5IF88 A0A0P5VR14 A0A0P5RKE3 A0A0P5RPT9 A0A0N8C7R8 A0A3B0J347 A0A0N8CE72 E9H736 A0A3B0J2M3 A0A0K8SQI6 A0A0A9ZGU3 A0A0P6CNX1 A0A182JIC1 A0A0A1X772 A0A0P6EJY1 A0A0Q9XLC5 B4KQ96 A0A0K8WJ51 A0A1L8I2T7 A0A1Q3G1V7 B4H8K7 B4J9Z7 P21271 P21271-3 Q28YI2 N6W4L9 A0A0T6BA34 A0A0Q9XHH8 F6WHW2 A0A182G783 A0A182RR43 Q02440 A0A1D5PRP3 A0A0P6BUJ9 A0A182V3H7 A0A182I464 G1PV90 A0A182WC09 A0A1B6DXB9 A0A2M4CPU8 A0A3Q0CRB3 G5E8G6 Q7Q5E6 G3X9Y9 A0A0L0CAC5
PDB
1W7J
E-value=0,
Score=1925
Ontologies
GO
GO:0005524
GO:0016459
GO:0051015
GO:0003774
GO:0003779
GO:0016021
GO:0005516
GO:0007291
GO:0043025
GO:0045856
GO:0006886
GO:0030899
GO:0047497
GO:0030048
GO:0032027
GO:0016028
GO:0042052
GO:0031475
GO:0030898
GO:0048471
GO:0099003
GO:0005903
GO:0050804
GO:0050775
GO:0098871
GO:0017137
GO:0098978
GO:1903078
GO:0032880
GO:0098944
GO:0016197
GO:0043005
GO:0045773
GO:0035255
GO:0015031
GO:0005886
GO:1903543
GO:0043197
GO:0032991
GO:0045179
GO:0016887
GO:0099159
GO:0016192
GO:0098685
GO:0060997
GO:0099639
GO:0008361
GO:0098856
GO:0048546
GO:0043588
GO:0061564
GO:0072659
GO:0032593
GO:0032869
GO:0060001
GO:0000146
GO:0031941
GO:0000139
GO:0005737
GO:0006396
GO:0000166
GO:0004887
GO:0003707
Topology
Subcellular location
Cytoplasm
Golgi apparatus membrane
Golgi apparatus membrane
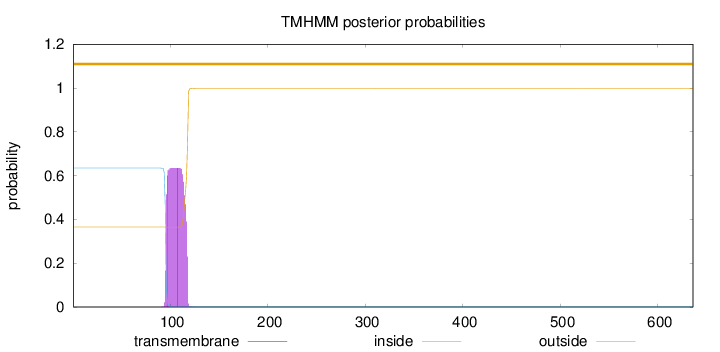
Length:
636
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.60193
Exp number, first 60 AAs:
0
Total prob of N-in:
0.63503
outside
1 - 636
Population Genetic Test Statistics
Pi
414.000504
Theta
185.101712
Tajima's D
2.723521
CLR
0.260582
CSRT
0.964551772411379
Interpretation
Possibly Balancing Selection
