Gene
KWMTBOMO00901 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009778
Annotation
PREDICTED:_LIM_and_SH3_domain_protein_Lasp_isoform_X4_[Papilio_polytes]
Full name
Nebulette
Alternative Name
Actin-binding Z-disk protein
Location in the cell
Nuclear Reliability : 3.34
Sequence
CDS
ATGGCCGAAACACCTGAATTGAAACGGATCGCCGAAAACACGAAACTACAGAGCAACGTGAAGTACCACGCCGACTTCGAGAGGAGTAAAGGGAAATTCACACAGGTGGCAGACGATCCTGAGACCTTAAGGATCAAAGCGAACACGAAGATCATAAGTAACGTGGCGTACCACGGCGATCTCGAGAAGAAAGCCCAGATGGAGAGGCAGAGGCAGATCAACGAAAACGGAGAAATAGTGGGTAGCAGTCATCAAACACAACATCAGCAGTCAAGGCAGATCGGTCGCGTAACCGATCTGGATCCGTTGGCCAATGAAGTGCAGAGACCGGCTAGCTCACAGAATCCGCAAGTGTACATAGCAATGTATGATTACGAAGCCAACGACAGTGATGAGGTGTCGTTCCGTGAAGGCGACCTCATAATGAATGTAAGTGCTATAGACGAGGGCTGGATGATGGGTCAGGTGGTACGCACCGGCATGGCGGGCATGCTGCCAGCCAACTACGTCGAGACCGCACCGGCGTCTGCATTACGGCAATACAATTGA
Protein
MAETPELKRIAENTKLQSNVKYHADFERSKGKFTQVADDPETLRIKANTKIISNVAYHGDLEKKAQMERQRQINENGEIVGSSHQTQHQQSRQIGRVTDLDPLANEVQRPASSQNPQVYIAMYDYEANDSDEVSFREGDLIMNVSAIDEGWMMGQVVRTGMAGMLPANYVETAPASALRQYN
Summary
Description
Binds to actin and plays an important role in the assembly of the Z-disk. May functionally link sarcomeric actin to the desmin intermediate filaments in the heart muscle sarcomeres (PubMed:27733623). Isoform 2 might play a role in the assembly of focal adhesion (PubMed:15004028).
Subunit
Isoform 2 interacts with ZYX (Zyxin) (PubMed:15004028). Interacts (via nebulin repeats 1-5) with DESM (via rod region) (PubMed:27733623).
Keywords
3D-structure
Actin-binding
Alternative splicing
Complete proteome
Cytoplasm
Methylation
Polymorphism
Reference proteome
Repeat
SH3 domain
Feature
chain Nebulette
splice variant In isoform 2.
sequence variant In dbSNP:rs75301590.
splice variant In isoform 2.
sequence variant In dbSNP:rs75301590.
Uniprot
A0A1E1WJR4
J3JX15
V5G4T0
A0A0J7NKB5
D6WRW3
A0A1W4XP20
+ More
A0A1W4XDQ4 A0A1Y1M1U2 A0A0Q9WT86 A0A0Q9XN10 A0A0K8TT92 A0A182MK35 M9NFH3 A0A1L8E5B0 A0A0R3P729 A0A1W7RAC5 A0A023GGL2 A0A224Z550 A0A1E1XL55 A0A131YRQ4 B7P377 A0A0B7BBX7 A0A091SZE2 A0A091QJ41 A0A146J2K6 A0A2I2UA04 A0A091TY58 A0A286ZNE6 B7ZB56 A0A341D4Q5 B4DH47 Q8N8M3 K9IIL1 A0A2K5ZHP0 A0A2R9CAM1 A0A287BFR4 A0A2I2ZD16 A0A2Y9KDC0 F1PQ88 A0A2U4B6T5 A0A340X7X9 A0A2Y9NYK7 A0A2U3VEE8 A0A384ARD3 A0A2K6FXT3 A0A287DEC2 O76041-2 A0A2K6BYR8 A0A2K6N6Q5 A0A2K5TRL9 A0A2I3FXH0 A0A2K5CX77 A0A2K6PT29 A0A2K5M6V8 A0A2J8RVW7 A0A2K5PCW0 A0A2K5J026 A0A2I3TGT5 U3C6Z0 A0A1D5RAK7 A0A1S3F0R1 A0A0P6K1Y3 A0A093ER58 A0A0A0AKK3 A0A2Y9RFS6 A0A091WCI3 A0A091V4G9 A0A087QUA3 A0A091PN56 A0A091J5N6 A0A093HUU3 A0A091F2D7 A0A094L6Q3 A0A093HMC4 A0A093I764 A0A091UUT3 A0A091QQ79
A0A1W4XDQ4 A0A1Y1M1U2 A0A0Q9WT86 A0A0Q9XN10 A0A0K8TT92 A0A182MK35 M9NFH3 A0A1L8E5B0 A0A0R3P729 A0A1W7RAC5 A0A023GGL2 A0A224Z550 A0A1E1XL55 A0A131YRQ4 B7P377 A0A0B7BBX7 A0A091SZE2 A0A091QJ41 A0A146J2K6 A0A2I2UA04 A0A091TY58 A0A286ZNE6 B7ZB56 A0A341D4Q5 B4DH47 Q8N8M3 K9IIL1 A0A2K5ZHP0 A0A2R9CAM1 A0A287BFR4 A0A2I2ZD16 A0A2Y9KDC0 F1PQ88 A0A2U4B6T5 A0A340X7X9 A0A2Y9NYK7 A0A2U3VEE8 A0A384ARD3 A0A2K6FXT3 A0A287DEC2 O76041-2 A0A2K6BYR8 A0A2K6N6Q5 A0A2K5TRL9 A0A2I3FXH0 A0A2K5CX77 A0A2K6PT29 A0A2K5M6V8 A0A2J8RVW7 A0A2K5PCW0 A0A2K5J026 A0A2I3TGT5 U3C6Z0 A0A1D5RAK7 A0A1S3F0R1 A0A0P6K1Y3 A0A093ER58 A0A0A0AKK3 A0A2Y9RFS6 A0A091WCI3 A0A091V4G9 A0A087QUA3 A0A091PN56 A0A091J5N6 A0A093HUU3 A0A091F2D7 A0A094L6Q3 A0A093HMC4 A0A093I764 A0A091UUT3 A0A091QQ79
Pubmed
22516182
23537049
18362917
19820115
28004739
17994087
+ More
26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 28797301 29209593 26830274 15632090 17975172 14702039 22722832 22398555 16341006 9733644 10470015 15004028 15164054 15489334 24129315 27733623 11140941 25362486 16136131 25243066 17431167 25319552
26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 28797301 29209593 26830274 15632090 17975172 14702039 22722832 22398555 16341006 9733644 10470015 15004028 15164054 15489334 24129315 27733623 11140941 25362486 16136131 25243066 17431167 25319552
EMBL
GDQN01003805
JAT87249.1
APGK01052202
BT127783
KB741211
KB632263
+ More
AEE62745.1 ENN72763.1 ERL90897.1 GALX01003426 JAB65040.1 LBMM01003986 KMQ92925.1 KQ971351 EFA06590.1 GEZM01042528 JAV79794.1 CH940647 KRF84335.1 CH933809 KRG06318.1 GDAI01000026 JAI17577.1 AXCM01000274 AE014296 AFH04470.1 GFDF01000163 JAV13921.1 CH379070 KRT08830.1 GFAH01000323 JAV48066.1 GBBM01002302 JAC33116.1 GFPF01010134 MAA21280.1 GFAA01003501 JAT99933.1 GEDV01007761 JAP80796.1 ABJB010418755 ABJB010491676 ABJB010776635 ABJB010892119 DS626813 EEC01049.1 HACG01042740 CEK89605.1 KK493376 KFQ64233.1 KK799327 KFQ27570.1 LC133191 CH473990 BAU79360.1 EDL78760.1 AANG04002256 KK467227 KFQ82525.1 AEMK02000074 AK316521 BAH14892.1 AK294926 BAG58008.1 AK096540 BAC04812.1 GABZ01007428 JAA46097.1 AJFE02018725 AJFE02018726 AJFE02018727 AJFE02018728 AJFE02018729 AJFE02018730 AJFE02018731 AJFE02018732 CABD030069448 CABD030069449 CABD030069450 CABD030069451 CABD030069452 CABD030069453 CABD030069454 CABD030069455 CABD030069456 CABD030069457 AAEX03001209 AGTP01041780 AGTP01041781 AGTP01041782 AGTP01041783 AGTP01041784 AGTP01041785 AGTP01041786 AGTP01041787 AGTP01041788 AGTP01041789 Y16241 Y17673 AF047368 AJ580772 AK295186 EF445000 AL359175 AL731547 AL157398 AL158160 CH471072 BC110453 BC126132 BC126134 AQIA01070259 AQIA01070260 ADFV01176518 ADFV01176519 ADFV01176520 ADFV01176521 ADFV01176522 ADFV01176523 ADFV01176524 ADFV01176525 ADFV01176526 ADFV01176527 NDHI03003644 PNJ12654.1 AACZ04070778 AACZ04070779 AACZ04070780 AACZ04070781 NBAG03000250 PNI61173.1 GAMT01008489 GAMS01001923 GAMR01006240 GAMP01005070 JAB03372.1 JAB21213.1 JAB27692.1 JAB47685.1 JSUE03043003 JSUE03043004 JSUE03043005 JU320560 AFE64316.1 GEBF01006026 JAN97606.1 KK368813 KFV42438.1 KL872299 KGL95024.1 KK735254 KFR13377.1 KL410573 KFQ97268.1 KL225912 KFM04807.1 KK661803 KFQ08683.1 KK501476 KFP15005.1 KK397951 KFV58288.1 KK719443 KFO63426.1 KL276503 KFZ66500.1 KL206260 KFV80615.1 KK586680 KFV97462.1 KL410182 KFQ94476.1 KK704005 KFQ29114.1
AEE62745.1 ENN72763.1 ERL90897.1 GALX01003426 JAB65040.1 LBMM01003986 KMQ92925.1 KQ971351 EFA06590.1 GEZM01042528 JAV79794.1 CH940647 KRF84335.1 CH933809 KRG06318.1 GDAI01000026 JAI17577.1 AXCM01000274 AE014296 AFH04470.1 GFDF01000163 JAV13921.1 CH379070 KRT08830.1 GFAH01000323 JAV48066.1 GBBM01002302 JAC33116.1 GFPF01010134 MAA21280.1 GFAA01003501 JAT99933.1 GEDV01007761 JAP80796.1 ABJB010418755 ABJB010491676 ABJB010776635 ABJB010892119 DS626813 EEC01049.1 HACG01042740 CEK89605.1 KK493376 KFQ64233.1 KK799327 KFQ27570.1 LC133191 CH473990 BAU79360.1 EDL78760.1 AANG04002256 KK467227 KFQ82525.1 AEMK02000074 AK316521 BAH14892.1 AK294926 BAG58008.1 AK096540 BAC04812.1 GABZ01007428 JAA46097.1 AJFE02018725 AJFE02018726 AJFE02018727 AJFE02018728 AJFE02018729 AJFE02018730 AJFE02018731 AJFE02018732 CABD030069448 CABD030069449 CABD030069450 CABD030069451 CABD030069452 CABD030069453 CABD030069454 CABD030069455 CABD030069456 CABD030069457 AAEX03001209 AGTP01041780 AGTP01041781 AGTP01041782 AGTP01041783 AGTP01041784 AGTP01041785 AGTP01041786 AGTP01041787 AGTP01041788 AGTP01041789 Y16241 Y17673 AF047368 AJ580772 AK295186 EF445000 AL359175 AL731547 AL157398 AL158160 CH471072 BC110453 BC126132 BC126134 AQIA01070259 AQIA01070260 ADFV01176518 ADFV01176519 ADFV01176520 ADFV01176521 ADFV01176522 ADFV01176523 ADFV01176524 ADFV01176525 ADFV01176526 ADFV01176527 NDHI03003644 PNJ12654.1 AACZ04070778 AACZ04070779 AACZ04070780 AACZ04070781 NBAG03000250 PNI61173.1 GAMT01008489 GAMS01001923 GAMR01006240 GAMP01005070 JAB03372.1 JAB21213.1 JAB27692.1 JAB47685.1 JSUE03043003 JSUE03043004 JSUE03043005 JU320560 AFE64316.1 GEBF01006026 JAN97606.1 KK368813 KFV42438.1 KL872299 KGL95024.1 KK735254 KFR13377.1 KL410573 KFQ97268.1 KL225912 KFM04807.1 KK661803 KFQ08683.1 KK501476 KFP15005.1 KK397951 KFV58288.1 KK719443 KFO63426.1 KL276503 KFZ66500.1 KL206260 KFV80615.1 KK586680 KFV97462.1 KL410182 KFQ94476.1 KK704005 KFQ29114.1
Proteomes
UP000019118
UP000030742
UP000036403
UP000007266
UP000192223
UP000008792
+ More
UP000009192 UP000075883 UP000000803 UP000001819 UP000001555 UP000011712 UP000008227 UP000252040 UP000233140 UP000240080 UP000001519 UP000248482 UP000002254 UP000245320 UP000265300 UP000248483 UP000245340 UP000261681 UP000233160 UP000005215 UP000005640 UP000233120 UP000233180 UP000233100 UP000001073 UP000233020 UP000233200 UP000233060 UP000233040 UP000233080 UP000002277 UP000008225 UP000006718 UP000081671 UP000053858 UP000248480 UP000053605 UP000053283 UP000053286 UP000053119 UP000052976 UP000053584
UP000009192 UP000075883 UP000000803 UP000001819 UP000001555 UP000011712 UP000008227 UP000252040 UP000233140 UP000240080 UP000001519 UP000248482 UP000002254 UP000245320 UP000265300 UP000248483 UP000245340 UP000261681 UP000233160 UP000005215 UP000005640 UP000233120 UP000233180 UP000233100 UP000001073 UP000233020 UP000233200 UP000233060 UP000233040 UP000233080 UP000002277 UP000008225 UP000006718 UP000081671 UP000053858 UP000248480 UP000053605 UP000053283 UP000053286 UP000053119 UP000052976 UP000053584
Interpro
SUPFAM
SSF50044
SSF50044
ProteinModelPortal
A0A1E1WJR4
J3JX15
V5G4T0
A0A0J7NKB5
D6WRW3
A0A1W4XP20
+ More
A0A1W4XDQ4 A0A1Y1M1U2 A0A0Q9WT86 A0A0Q9XN10 A0A0K8TT92 A0A182MK35 M9NFH3 A0A1L8E5B0 A0A0R3P729 A0A1W7RAC5 A0A023GGL2 A0A224Z550 A0A1E1XL55 A0A131YRQ4 B7P377 A0A0B7BBX7 A0A091SZE2 A0A091QJ41 A0A146J2K6 A0A2I2UA04 A0A091TY58 A0A286ZNE6 B7ZB56 A0A341D4Q5 B4DH47 Q8N8M3 K9IIL1 A0A2K5ZHP0 A0A2R9CAM1 A0A287BFR4 A0A2I2ZD16 A0A2Y9KDC0 F1PQ88 A0A2U4B6T5 A0A340X7X9 A0A2Y9NYK7 A0A2U3VEE8 A0A384ARD3 A0A2K6FXT3 A0A287DEC2 O76041-2 A0A2K6BYR8 A0A2K6N6Q5 A0A2K5TRL9 A0A2I3FXH0 A0A2K5CX77 A0A2K6PT29 A0A2K5M6V8 A0A2J8RVW7 A0A2K5PCW0 A0A2K5J026 A0A2I3TGT5 U3C6Z0 A0A1D5RAK7 A0A1S3F0R1 A0A0P6K1Y3 A0A093ER58 A0A0A0AKK3 A0A2Y9RFS6 A0A091WCI3 A0A091V4G9 A0A087QUA3 A0A091PN56 A0A091J5N6 A0A093HUU3 A0A091F2D7 A0A094L6Q3 A0A093HMC4 A0A093I764 A0A091UUT3 A0A091QQ79
A0A1W4XDQ4 A0A1Y1M1U2 A0A0Q9WT86 A0A0Q9XN10 A0A0K8TT92 A0A182MK35 M9NFH3 A0A1L8E5B0 A0A0R3P729 A0A1W7RAC5 A0A023GGL2 A0A224Z550 A0A1E1XL55 A0A131YRQ4 B7P377 A0A0B7BBX7 A0A091SZE2 A0A091QJ41 A0A146J2K6 A0A2I2UA04 A0A091TY58 A0A286ZNE6 B7ZB56 A0A341D4Q5 B4DH47 Q8N8M3 K9IIL1 A0A2K5ZHP0 A0A2R9CAM1 A0A287BFR4 A0A2I2ZD16 A0A2Y9KDC0 F1PQ88 A0A2U4B6T5 A0A340X7X9 A0A2Y9NYK7 A0A2U3VEE8 A0A384ARD3 A0A2K6FXT3 A0A287DEC2 O76041-2 A0A2K6BYR8 A0A2K6N6Q5 A0A2K5TRL9 A0A2I3FXH0 A0A2K5CX77 A0A2K6PT29 A0A2K5M6V8 A0A2J8RVW7 A0A2K5PCW0 A0A2K5J026 A0A2I3TGT5 U3C6Z0 A0A1D5RAK7 A0A1S3F0R1 A0A0P6K1Y3 A0A093ER58 A0A0A0AKK3 A0A2Y9RFS6 A0A091WCI3 A0A091V4G9 A0A087QUA3 A0A091PN56 A0A091J5N6 A0A093HUU3 A0A091F2D7 A0A094L6Q3 A0A093HMC4 A0A093I764 A0A091UUT3 A0A091QQ79
PDB
1NEB
E-value=6.8715e-16,
Score=200
Ontologies
GO
Topology
Subcellular location
Cytoplasm
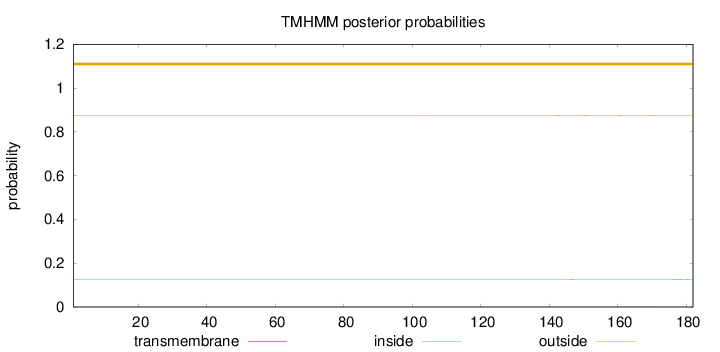
Length:
182
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0316
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12483
outside
1 - 182
Population Genetic Test Statistics
Pi
269.963633
Theta
179.662282
Tajima's D
1.968049
CLR
0.113997
CSRT
0.877706114694265
Interpretation
Uncertain
