Gene
KWMTBOMO00893
Pre Gene Modal
BGIBMGA009658
Annotation
PREDICTED:_WD_repeat-containing_protein_7_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.469
Sequence
CDS
ATGGGTCCCGGTAACCGCTTAATTCCTTGTCGTCCAAGGAAGGATAAACGGCGTAAGACCGCGCGTGAGGTCTCCCATGCTGCGGACCAACCAAAATGGGTGCAATTGGACGTCCTGGAGCAGGATGATAAGGCGGGGCATTATGACGGTACAACTCCGAAGAAGCGTCCCAAAGAGATGTGTCTCTCAGACGCTCCCCACACAATGGAGATCGCCCAGATACTTGTGTCGCTGCTGCACGCCTGGGGTCTCGACCCTGACCTGGACAGGGTCTGCGAGTCGAAGCTCGGTCTATTAAGACCTATGGTCCCGGTGTGCTTCGGCGTGGTCTCCCGACACGGCCACATGGCTGTCCAGCTTCCGACTCGCCTCATCAGCTGGACGGAGGCCTGCGCTGCCGCGCCCGACGACCCGCTACTCGTAACCTCGGATCTACCACCGGAACTATTGACCCAGGAAAAACTTACCAGGCTGTTCTCTTCGAAGCTACACTGGGAGTTGAGCACGACGCTGACCACGAACCATTTGCTGTCCATCGTGGCCTTAGCCAACACCCTCATGTCGATGAGCAACGCCAGCTTCCTACCAGAATCGGAGATGGCGCGGAGATTGCACAGACCTTCGACCCGAAGCTCTTCTTCATGGGCGGCGGAGGAAGAACGTGAGGAACAGCTAGCAGCCCAGCAGGCCCACATCAAGCAGGGCTGGTCGTTACTCGCCACGCTCCATTGCGTATTGCTCCCTGACAAAGTCGTTGCAACCGGAGCGAAGAACTTCAAGCGTCCCCAAGTGGAGATGCTGGCGCGCAGGTGGCAGCATCACTGTCTGGAAGTGAGGGAGGCAGCCCAGGCTCTACTCCTAGCTGAATTAGCAAGGTTGGGTCCGAAGGGTCGCAAAGCCCTGGTCGACAACTGGGCCCAGTATCTACCTCTGTACACGCACACAGAGAGCATCAACCCTCATGCGGCGCAGAAGGAACCGGCCGAGAAATTGGCCAAAACGGTCGATGAGTTTGACGAGGAAGAAGAAGAGATAGTCCGGAAGCCATCTTCCCTCGCCGAGCTGAAGCGGAAACAGACAACGGCCGTGGTCCTGCTCGGAGTCATAGGGGCGGAGTTCGGACAGGACATCACAAGCGAAGGTGCGGCCGGGAACAAGCGCCGTAAGGATGTCGACAGCAGGAAGAGTTCGGTTGTAGAAGGATTCACGCTAAGCTCTTCGAATAACTTGGCCCGTCTGACTGCACTGGCCCTGACGCACTTACTGCAGGCGCCCGCGTGTCCCCGCCTGCCAGCCCACACGCCCCTGCGGCGTGCCGCTGTTGATCTGCTGGGGAGGGGCTTCACAGTTTGGGAACCATATTTGGATGTCAGTCACGTTCTTTTAGGTTTGCTGGAGATGTGTTCGGACGCGGATAAGTTGGTTCCGTCGATGACGTACGGTCTCCCGCTGACCGCGCAGGCCGACTCGTGTCGCACGGCGCGACACGCGCTGACACTCATCGCCACCGCCAGGCCGGCGGCGTTCATAACAACGATGGCCCGCGAGGTGGCGCGGTGCGCGGCCGCGGCCCAGAGCGGACAGCCCTCCCCGGCGTACCACGCGCTACAGAAGGGCAGGGCCGAGGTGCTCCGCGGGATCGAGCTGCTCATTGAGAAGATGCACAGCGAGGTCGCCGAACTGCTTGTCGAGGTGATGGATATAATCCTGCACTGCGTGGACCAGTCGCACCTGAAGAGTAAGGGTTTGAACGAGGTGTTCCCGGCCATCTGTCGGTACAACCAGGTCTCGCACTGCCCAGCAACCCGGAGAATTGCGGTCGGCAGCCACACTGGGCAGCTAGCGCTATACGAGCTGCGGGCGGGGCGCTGCCAGTCCCTGGCCGCACACAGCGGGCCCGTCACCGCCTGCGCCTTCAGCCCCGACGGCAGATACCTGGTCTCCTACGCCACGACCGACAACCGACTCTCCTTCTGGCAGAGCACCTCTGGTATGTTCGGTCTGGGCGCGGCGCAGACCCGCTGCGTGAAGTGCTACAGCACGGCGCCCATGGCGGACGTGGCGCGGCTGAACCCGGCCCGCCTGGCCCGCCTCGTGTGGACCAACTCCAGGACCGTCACCCTCATGCTGGCCGACGGCTCCGAAACCCGGTTCAATGTTTGA
Protein
MGPGNRLIPCRPRKDKRRKTAREVSHAADQPKWVQLDVLEQDDKAGHYDGTTPKKRPKEMCLSDAPHTMEIAQILVSLLHAWGLDPDLDRVCESKLGLLRPMVPVCFGVVSRHGHMAVQLPTRLISWTEACAAAPDDPLLVTSDLPPELLTQEKLTRLFSSKLHWELSTTLTTNHLLSIVALANTLMSMSNASFLPESEMARRLHRPSTRSSSSWAAEEEREEQLAAQQAHIKQGWSLLATLHCVLLPDKVVATGAKNFKRPQVEMLARRWQHHCLEVREAAQALLLAELARLGPKGRKALVDNWAQYLPLYTHTESINPHAAQKEPAEKLAKTVDEFDEEEEEIVRKPSSLAELKRKQTTAVVLLGVIGAEFGQDITSEGAAGNKRRKDVDSRKSSVVEGFTLSSSNNLARLTALALTHLLQAPACPRLPAHTPLRRAAVDLLGRGFTVWEPYLDVSHVLLGLLEMCSDADKLVPSMTYGLPLTAQADSCRTARHALTLIATARPAAFITTMAREVARCAAAAQSGQPSPAYHALQKGRAEVLRGIELLIEKMHSEVAELLVEVMDIILHCVDQSHLKSKGLNEVFPAICRYNQVSHCPATRRIAVGSHTGQLALYELRAGRCQSLAAHSGPVTACAFSPDGRYLVSYATTDNRLSFWQSTSGMFGLGAAQTRCVKCYSTAPMADVARLNPARLARLVWTNSRTVTLMLADGSETRFNV
Summary
Similarity
Belongs to the cyclin family.
Uniprot
H9JJF7
A0A2A4IWL8
A0A2A4IV76
A0A194R0H3
A0A194PZL5
D6WQ49
+ More
A0A1Y1K2X3 A0A1Y1K5S4 A0A1Y1K5X2 A0A1Y1K6H9 A0A026WNU8 U4ULR5 E2A854 A0A0J7KMC8 E2B7B4 A0A0C9R6H5 A0A154NX68 A0A158NPH6 F4WP01 A0A088ATB4 A0A2A3EI53 A0A0M8ZT94 A0A310SFQ4 A0A0L7R9M4 A0A1B6LRM6 A0A1B6LFK9 A0A232F2T0 K7IUY9 A0A1B6CTQ7 A0A1B6DM46 A0A1B6C9P0 A0A1B6CWJ7 A0A1B6FXU2 A0A182T8V9 A0A1B6KFE9 A0A182MD63 A0A2M4A0H3 A0A2M4CTS9 A0A2M4B982 A0A2M4B9A8 A0A182FWH2 A0A1B6MPX3 A0A1B6LGD1 A0A182J8U4 A0A182RFI1 A0A182W7E0 A0A084WD89 Q5TQM2 A0A1S4GWY3 A0A067QWM1 A0A182TLG9 A0A182JU60 A0A182L1V5 A0A182V462 A0A182HV02 A0A182WT05 A0A1B6KPF7 A0A182NTN2 A0A1Q3FHD3 A0A1Q3FH64 A0A182Q0N8 B0W6Z8 A0A1S4J7T9 A0A1Q3FH77 A0A1Q3FH73 A0A023F514 A0A069DXN0 A0A0P4VWE9 A0A151WRU5 A0A195B974 A0A195EUZ3 A0A224XGA0 A0A195CP23 A0A195E576 A0A182H3E7 A0A1L8DN80 Q16IV4 A0A034WAF8 W8AI12 A0A1L8DNJ5 A0A0K8U841 A0A0A1XCN5 A0A0L0CTE4 A0A0M4ETH8 A0A182P4S5 T1PE41 A0A1B0FNG5 A0A0T6BA33 A0A1I8MI76 T1IGI3 A0A1A9WGD5 A0A1I8NT69 A0A0K8T9P9 A0A0K8T9A7 A0A0A9XHP6 N6T562 A0A182XZY3 E0VAC8
A0A1Y1K2X3 A0A1Y1K5S4 A0A1Y1K5X2 A0A1Y1K6H9 A0A026WNU8 U4ULR5 E2A854 A0A0J7KMC8 E2B7B4 A0A0C9R6H5 A0A154NX68 A0A158NPH6 F4WP01 A0A088ATB4 A0A2A3EI53 A0A0M8ZT94 A0A310SFQ4 A0A0L7R9M4 A0A1B6LRM6 A0A1B6LFK9 A0A232F2T0 K7IUY9 A0A1B6CTQ7 A0A1B6DM46 A0A1B6C9P0 A0A1B6CWJ7 A0A1B6FXU2 A0A182T8V9 A0A1B6KFE9 A0A182MD63 A0A2M4A0H3 A0A2M4CTS9 A0A2M4B982 A0A2M4B9A8 A0A182FWH2 A0A1B6MPX3 A0A1B6LGD1 A0A182J8U4 A0A182RFI1 A0A182W7E0 A0A084WD89 Q5TQM2 A0A1S4GWY3 A0A067QWM1 A0A182TLG9 A0A182JU60 A0A182L1V5 A0A182V462 A0A182HV02 A0A182WT05 A0A1B6KPF7 A0A182NTN2 A0A1Q3FHD3 A0A1Q3FH64 A0A182Q0N8 B0W6Z8 A0A1S4J7T9 A0A1Q3FH77 A0A1Q3FH73 A0A023F514 A0A069DXN0 A0A0P4VWE9 A0A151WRU5 A0A195B974 A0A195EUZ3 A0A224XGA0 A0A195CP23 A0A195E576 A0A182H3E7 A0A1L8DN80 Q16IV4 A0A034WAF8 W8AI12 A0A1L8DNJ5 A0A0K8U841 A0A0A1XCN5 A0A0L0CTE4 A0A0M4ETH8 A0A182P4S5 T1PE41 A0A1B0FNG5 A0A0T6BA33 A0A1I8MI76 T1IGI3 A0A1A9WGD5 A0A1I8NT69 A0A0K8T9P9 A0A0K8T9A7 A0A0A9XHP6 N6T562 A0A182XZY3 E0VAC8
Pubmed
EMBL
BABH01017727
BABH01017728
NWSH01006075
PCG63684.1
PCG63685.1
KQ460890
+ More
KPJ11024.1 KQ459593 KPI96605.1 KQ971354 EFA06130.2 GEZM01094193 JAV55819.1 GEZM01094191 JAV55821.1 GEZM01094194 JAV55818.1 GEZM01094192 JAV55820.1 KK107144 EZA57603.1 KB632263 ERL90940.1 GL437497 EFN70366.1 LBMM01005603 KMQ91391.1 GL446154 EFN88399.1 GBYB01002366 GBYB01012661 JAG72133.1 JAG82428.1 KQ434777 KZC04217.1 ADTU01022546 ADTU01022547 GL888243 EGI63932.1 KZ288253 PBC30952.1 KQ435853 KOX70670.1 KQ761797 OAD56789.1 KQ414621 KOC67575.1 GEBQ01013672 JAT26305.1 GEBQ01017501 JAT22476.1 NNAY01001187 OXU24819.1 AAZX01004672 GEDC01020537 JAS16761.1 GEDC01010608 JAS26690.1 GEDC01027127 JAS10171.1 GEDC01019399 JAS17899.1 GECZ01014757 JAS55012.1 GEBQ01029832 JAT10145.1 AXCM01003107 GGFK01000931 MBW34252.1 GGFL01004501 MBW68679.1 GGFJ01000473 MBW49614.1 GGFJ01000472 MBW49613.1 GEBQ01001981 JAT37996.1 GEBQ01017204 JAT22773.1 ATLV01022956 KE525338 KFB48183.1 AAAB01008964 EAL39674.3 KK853162 KDR10431.1 APCN01002488 GEBQ01026639 JAT13338.1 GFDL01008132 JAV26913.1 GFDL01008149 JAV26896.1 AXCN02000795 DS231851 EDS37272.1 GFDL01008138 JAV26907.1 GFDL01008160 JAV26885.1 GBBI01002588 JAC16124.1 GBGD01000096 JAC88793.1 GDKW01001577 JAI55018.1 KQ982807 KYQ50395.1 KQ976542 KYM81086.1 KQ981965 KYN31712.1 GFTR01008814 JAW07612.1 KQ977574 KYN01859.1 KQ979608 KYN20340.1 JXUM01107375 JXUM01107376 JXUM01107377 JXUM01107378 KQ565136 KXJ71294.1 GFDF01006161 JAV07923.1 CH478049 EAT34201.1 GAKP01008194 JAC50758.1 GAMC01018420 GAMC01018419 JAB88135.1 GFDF01006137 JAV07947.1 GDHF01029487 JAI22827.1 GBXI01006069 JAD08223.1 JRES01000037 KNC34669.1 CP012528 ALC49827.1 KA646991 AFP61620.1 CCAG010019210 LJIG01002744 KRT84196.1 ACPB03001109 GBRD01003710 JAG62111.1 GBRD01003712 JAG62109.1 GBHO01024086 GBHO01024085 GBHO01024084 GBHO01024082 JAG19518.1 JAG19519.1 JAG19520.1 JAG19522.1 APGK01052253 APGK01052254 KB741211 ENN72803.1 DS235005 EEB10334.1
KPJ11024.1 KQ459593 KPI96605.1 KQ971354 EFA06130.2 GEZM01094193 JAV55819.1 GEZM01094191 JAV55821.1 GEZM01094194 JAV55818.1 GEZM01094192 JAV55820.1 KK107144 EZA57603.1 KB632263 ERL90940.1 GL437497 EFN70366.1 LBMM01005603 KMQ91391.1 GL446154 EFN88399.1 GBYB01002366 GBYB01012661 JAG72133.1 JAG82428.1 KQ434777 KZC04217.1 ADTU01022546 ADTU01022547 GL888243 EGI63932.1 KZ288253 PBC30952.1 KQ435853 KOX70670.1 KQ761797 OAD56789.1 KQ414621 KOC67575.1 GEBQ01013672 JAT26305.1 GEBQ01017501 JAT22476.1 NNAY01001187 OXU24819.1 AAZX01004672 GEDC01020537 JAS16761.1 GEDC01010608 JAS26690.1 GEDC01027127 JAS10171.1 GEDC01019399 JAS17899.1 GECZ01014757 JAS55012.1 GEBQ01029832 JAT10145.1 AXCM01003107 GGFK01000931 MBW34252.1 GGFL01004501 MBW68679.1 GGFJ01000473 MBW49614.1 GGFJ01000472 MBW49613.1 GEBQ01001981 JAT37996.1 GEBQ01017204 JAT22773.1 ATLV01022956 KE525338 KFB48183.1 AAAB01008964 EAL39674.3 KK853162 KDR10431.1 APCN01002488 GEBQ01026639 JAT13338.1 GFDL01008132 JAV26913.1 GFDL01008149 JAV26896.1 AXCN02000795 DS231851 EDS37272.1 GFDL01008138 JAV26907.1 GFDL01008160 JAV26885.1 GBBI01002588 JAC16124.1 GBGD01000096 JAC88793.1 GDKW01001577 JAI55018.1 KQ982807 KYQ50395.1 KQ976542 KYM81086.1 KQ981965 KYN31712.1 GFTR01008814 JAW07612.1 KQ977574 KYN01859.1 KQ979608 KYN20340.1 JXUM01107375 JXUM01107376 JXUM01107377 JXUM01107378 KQ565136 KXJ71294.1 GFDF01006161 JAV07923.1 CH478049 EAT34201.1 GAKP01008194 JAC50758.1 GAMC01018420 GAMC01018419 JAB88135.1 GFDF01006137 JAV07947.1 GDHF01029487 JAI22827.1 GBXI01006069 JAD08223.1 JRES01000037 KNC34669.1 CP012528 ALC49827.1 KA646991 AFP61620.1 CCAG010019210 LJIG01002744 KRT84196.1 ACPB03001109 GBRD01003710 JAG62111.1 GBRD01003712 JAG62109.1 GBHO01024086 GBHO01024085 GBHO01024084 GBHO01024082 JAG19518.1 JAG19519.1 JAG19520.1 JAG19522.1 APGK01052253 APGK01052254 KB741211 ENN72803.1 DS235005 EEB10334.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007266
UP000053097
+ More
UP000030742 UP000000311 UP000036403 UP000008237 UP000076502 UP000005205 UP000007755 UP000005203 UP000242457 UP000053105 UP000053825 UP000215335 UP000002358 UP000075901 UP000075883 UP000069272 UP000075880 UP000075900 UP000075920 UP000030765 UP000007062 UP000027135 UP000075902 UP000075881 UP000075882 UP000075903 UP000075840 UP000076407 UP000075884 UP000075886 UP000002320 UP000075809 UP000078540 UP000078541 UP000078542 UP000078492 UP000069940 UP000249989 UP000008820 UP000037069 UP000092553 UP000075885 UP000092444 UP000095301 UP000015103 UP000091820 UP000095300 UP000019118 UP000076408 UP000009046
UP000030742 UP000000311 UP000036403 UP000008237 UP000076502 UP000005205 UP000007755 UP000005203 UP000242457 UP000053105 UP000053825 UP000215335 UP000002358 UP000075901 UP000075883 UP000069272 UP000075880 UP000075900 UP000075920 UP000030765 UP000007062 UP000027135 UP000075902 UP000075881 UP000075882 UP000075903 UP000075840 UP000076407 UP000075884 UP000075886 UP000002320 UP000075809 UP000078540 UP000078541 UP000078542 UP000078492 UP000069940 UP000249989 UP000008820 UP000037069 UP000092553 UP000075885 UP000092444 UP000095301 UP000015103 UP000091820 UP000095300 UP000019118 UP000076408 UP000009046
PRIDE
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR019775 WD40_repeat_CS
IPR011041 Quinoprot_gluc/sorb_DH
IPR036915 Cyclin-like_sf
IPR013763 Cyclin-like
IPR006671 Cyclin_N
IPR004367 Cyclin_C-dom
IPR011044 Quino_amine_DH_bsu
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR019775 WD40_repeat_CS
IPR011041 Quinoprot_gluc/sorb_DH
IPR036915 Cyclin-like_sf
IPR013763 Cyclin-like
IPR006671 Cyclin_N
IPR004367 Cyclin_C-dom
IPR011044 Quino_amine_DH_bsu
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JJF7
A0A2A4IWL8
A0A2A4IV76
A0A194R0H3
A0A194PZL5
D6WQ49
+ More
A0A1Y1K2X3 A0A1Y1K5S4 A0A1Y1K5X2 A0A1Y1K6H9 A0A026WNU8 U4ULR5 E2A854 A0A0J7KMC8 E2B7B4 A0A0C9R6H5 A0A154NX68 A0A158NPH6 F4WP01 A0A088ATB4 A0A2A3EI53 A0A0M8ZT94 A0A310SFQ4 A0A0L7R9M4 A0A1B6LRM6 A0A1B6LFK9 A0A232F2T0 K7IUY9 A0A1B6CTQ7 A0A1B6DM46 A0A1B6C9P0 A0A1B6CWJ7 A0A1B6FXU2 A0A182T8V9 A0A1B6KFE9 A0A182MD63 A0A2M4A0H3 A0A2M4CTS9 A0A2M4B982 A0A2M4B9A8 A0A182FWH2 A0A1B6MPX3 A0A1B6LGD1 A0A182J8U4 A0A182RFI1 A0A182W7E0 A0A084WD89 Q5TQM2 A0A1S4GWY3 A0A067QWM1 A0A182TLG9 A0A182JU60 A0A182L1V5 A0A182V462 A0A182HV02 A0A182WT05 A0A1B6KPF7 A0A182NTN2 A0A1Q3FHD3 A0A1Q3FH64 A0A182Q0N8 B0W6Z8 A0A1S4J7T9 A0A1Q3FH77 A0A1Q3FH73 A0A023F514 A0A069DXN0 A0A0P4VWE9 A0A151WRU5 A0A195B974 A0A195EUZ3 A0A224XGA0 A0A195CP23 A0A195E576 A0A182H3E7 A0A1L8DN80 Q16IV4 A0A034WAF8 W8AI12 A0A1L8DNJ5 A0A0K8U841 A0A0A1XCN5 A0A0L0CTE4 A0A0M4ETH8 A0A182P4S5 T1PE41 A0A1B0FNG5 A0A0T6BA33 A0A1I8MI76 T1IGI3 A0A1A9WGD5 A0A1I8NT69 A0A0K8T9P9 A0A0K8T9A7 A0A0A9XHP6 N6T562 A0A182XZY3 E0VAC8
A0A1Y1K2X3 A0A1Y1K5S4 A0A1Y1K5X2 A0A1Y1K6H9 A0A026WNU8 U4ULR5 E2A854 A0A0J7KMC8 E2B7B4 A0A0C9R6H5 A0A154NX68 A0A158NPH6 F4WP01 A0A088ATB4 A0A2A3EI53 A0A0M8ZT94 A0A310SFQ4 A0A0L7R9M4 A0A1B6LRM6 A0A1B6LFK9 A0A232F2T0 K7IUY9 A0A1B6CTQ7 A0A1B6DM46 A0A1B6C9P0 A0A1B6CWJ7 A0A1B6FXU2 A0A182T8V9 A0A1B6KFE9 A0A182MD63 A0A2M4A0H3 A0A2M4CTS9 A0A2M4B982 A0A2M4B9A8 A0A182FWH2 A0A1B6MPX3 A0A1B6LGD1 A0A182J8U4 A0A182RFI1 A0A182W7E0 A0A084WD89 Q5TQM2 A0A1S4GWY3 A0A067QWM1 A0A182TLG9 A0A182JU60 A0A182L1V5 A0A182V462 A0A182HV02 A0A182WT05 A0A1B6KPF7 A0A182NTN2 A0A1Q3FHD3 A0A1Q3FH64 A0A182Q0N8 B0W6Z8 A0A1S4J7T9 A0A1Q3FH77 A0A1Q3FH73 A0A023F514 A0A069DXN0 A0A0P4VWE9 A0A151WRU5 A0A195B974 A0A195EUZ3 A0A224XGA0 A0A195CP23 A0A195E576 A0A182H3E7 A0A1L8DN80 Q16IV4 A0A034WAF8 W8AI12 A0A1L8DNJ5 A0A0K8U841 A0A0A1XCN5 A0A0L0CTE4 A0A0M4ETH8 A0A182P4S5 T1PE41 A0A1B0FNG5 A0A0T6BA33 A0A1I8MI76 T1IGI3 A0A1A9WGD5 A0A1I8NT69 A0A0K8T9P9 A0A0K8T9A7 A0A0A9XHP6 N6T562 A0A182XZY3 E0VAC8
PDB
4CY5
E-value=0.000228366,
Score=108
Ontologies
GO
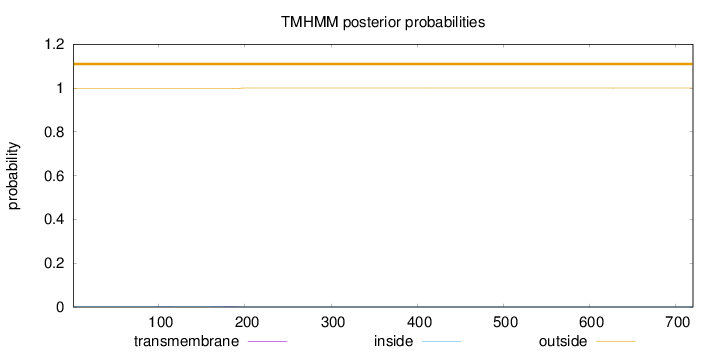
Topology
Length:
720
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0467700000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00198
outside
1 - 720
Population Genetic Test Statistics
Pi
239.477678
Theta
150.177519
Tajima's D
1.873688
CLR
0.382927
CSRT
0.859957002149892
Interpretation
Uncertain
