Gene
KWMTBOMO00889
Pre Gene Modal
BGIBMGA009661
Annotation
PREDICTED:_uncharacterized_protein_LOC101737750_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.418
Sequence
CDS
ATGTTTTATTTTTCTTTATTAGATATTGTTAAAGAAAATATACGTAACTATGAAATCATAGGTAGTGGAAGTACTCCTGGCTGGCTATTGGTTCCACTCAAACAAAATCCATATCATGTTTCTATTATTTCCAGTGACCTCGCTGATGAGGAAGATAAGAAGTCTCCGGTGCTCCTTTTAGGGGTGAATAAGATCTCCGACTCGCCGTACGTAAGGAGGAGATCGATAGAACCTCATTCGTCGGGCTCTATAAGATTCTTCAGACCGCCCAAGTCCAAATACACAAGGGGCCAGCACGGTGATGAGAGACTGGAACAGATAAGGAAGGACCTCCTCGTTCACAAGAATAAACCGGACAGCTACATTCCGAAGGTCGAGCACGAGATCTTCGTTGAGCAGGAGGAGAAGTGCTCAAAGCTGAGCTGCGTCGGCAATTGCATCTGCGATCGGAAGAAGAGCTCCAATCGCGGCTCTTTCTCGTTCCACAGACCCGACCAACTCCTTCCCAATTACCAACCACAATGTAGGGACTACTCTGAGAACGAGATAGAATACAAATACACAGAACACATTCCACTAAACCAGCCGTCGAGTAGTAAGCAGAGTTATAACGTCACCCCTAAATCAACGGAAAGAAGACACTTCCTACTCGCGAACCTTAACGAGAGATTCAGAAAGACTCTGAGCTTGGACTCCCGCAAGGACACGAGGGTGCTAAGTCTGCCCCAAGAAAACCGACCTAAATCATTCATAAATACGAGCAACATTTGCGTGCAACAATTCAAATCGGGCGACCCCTTCATCCCGACGAATTTTAACAAGGGCAGCCAACCTAAGAAAAAGCGGAAGTCGTTTTTTTCGCTAGACACGTTTTTCGATACGAAAAAGTCGGACACGTCATCCGTTGACGATTTTTGCTCGTCCAAATATAAAAGATTCGAACTCGAGAGCGATGACTCGCCTGTATTCAGACGCGACGATCACAAACGGGTGAAGACGCCCGATTTGCTAAACCTACCGGTCATCATTGATTCGGTTAAGAAAAAGCATTCCGAAACACGAAGACAGCTGGAGAAGCTAGAGAACTATTACGTGAAAAACTTAAATAAAACCCAAGCGGTCGTAGATGCGGTTCCGAAGGACGCAACCGATGTCCAAGGGCCGAGCGGACACGCCAATCACGTCTACATAGACGACGAGGACGTGGAATACATTGAGCCGATACGCAGCAACTTCACAAGCCAAAGCACTTTGATCCTGGACAAAAACATTCCTATACCAGACCCCGCAAATATATCCGATGCCGAAAATCATATAACCGTATCACATAATGACAGAAATGACATTGACAGTATAGGACATTACGAAATCGAAGTTAAGAAATCAGATTTAATAAAGAATCGGGTCGCGGATTTAATTGTTAAAGTGATGGATAAGTCGGTGGATTCTATAGGTAGTTGTTCGTTGGACGTGGATGCCAGCACGGATTTCTCAGATACAACATCGGGTAGTCTGAATCTCCTAACTCCATCGTCTACCACAAGTCGAGTGAGAGACTTCACATCCCGCATACAAGAGCGGGCCGGCTCGAATCTACAACAAAACTACTTGACCCCAACCCCGGTGGTCACCCCAATCAGGACCCCAGAGGAGACCACACCGACTCCTAGGAAATCTGAGCATCTCCTGAAACCACCCCCAAAAGTTCACGTCGACTCGTCAAGTCATCGATCCCGTGGACAACACAAACGTAGAGACGACCTGCCCCAAAAGAAACCAAGCTACTTGAACCTAGCCTGTTCCGTAAATGGCTATACCAACTTAACGACCTACGACTCGAAACTAAGGCAAGACATAAACAAGAGCCGGGAAGCGTCACCAATCCGACCGATAACATACACATATCAGTATAGAAGCGAAAATAATTCACTCTTGGTACCCGTGCCGGTCAGCGCGAACAAACTGCTCGTCCCCACCTTCGGGCCTCATGATACAACCGATTTGACGGCGAAAGCGCCTTCCAAGGCGCACACTGATCCGTCGCTGGCAACATCGACCCTCCATGTGTATATGGCCGCTGAAAACAAACAGTTGAAAGACGATTTTCTGGGCCCCGACATGACGACCACAAGCCGCCCTTACATATCGACAGAGAGCAAGAACTTTGCTACCTCGATGCTCCATCAGAAAGACGAATTGGACAATGCAAAGGAGATCACCTTCAAATCGAGCTATTCCGAGACGAATTTCCGGAAGACTGTGACCAGTAACGGCAAGGACTCTAGGTTTACATCGGAATCTTACACGATATCGTCGAACGGGATGTCGAAGAGGGTCGAAATAACGAAAGAGAATGGAGAAAAGTTGACGAGTCCCATGAAAAGCTTCATTCAACAACGGGTCGAGAGGTTGTACGGCCCCGGAGCTTTAGCCCAGGGGTTTTTTAACCAGAAAAGATTGAAAAGTCAAAGTGAAGACGATGATTCGAAGGTATTGACAGAGAAATCTCTAAATTGTCCGGAATACTCGGTAAGGAAACTAAACAAGAGCTTTGATGACGCCTACACTAGTCCAAATGGGAATTTAGATTCTAGCGTCTCTCTTCCTGTGCTCAGGCACCTGAGACCCGAATTTCGGGCCCAATTACCAATACATTCACCAAAAAAGAGTGTTAAAGCTGAACTATCGCCGCAGAAATTAGAAGAAGACATTCCACGACTGGACACGAAGACTGAACTCAGCGAAGTGGTGACGAACGGTGTATCTGTGATAAAAATTGATGATTCCTTGAATAACGATGCTGTCAACGGCAAGGAAATCAATGGGGAGGCAGTCAAAGATGCCTATTACTTTTTGGATCTCGAGAAGAAGGAGACTGAGAGGTTATTGGCATTGTCTGCCAGTGCTGAAAAGGAATTGGAGAATTTACAGGTAATTACAACTTTTACATTTTAA
Protein
MFYFSLLDIVKENIRNYEIIGSGSTPGWLLVPLKQNPYHVSIISSDLADEEDKKSPVLLLGVNKISDSPYVRRRSIEPHSSGSIRFFRPPKSKYTRGQHGDERLEQIRKDLLVHKNKPDSYIPKVEHEIFVEQEEKCSKLSCVGNCICDRKKSSNRGSFSFHRPDQLLPNYQPQCRDYSENEIEYKYTEHIPLNQPSSSKQSYNVTPKSTERRHFLLANLNERFRKTLSLDSRKDTRVLSLPQENRPKSFINTSNICVQQFKSGDPFIPTNFNKGSQPKKKRKSFFSLDTFFDTKKSDTSSVDDFCSSKYKRFELESDDSPVFRRDDHKRVKTPDLLNLPVIIDSVKKKHSETRRQLEKLENYYVKNLNKTQAVVDAVPKDATDVQGPSGHANHVYIDDEDVEYIEPIRSNFTSQSTLILDKNIPIPDPANISDAENHITVSHNDRNDIDSIGHYEIEVKKSDLIKNRVADLIVKVMDKSVDSIGSCSLDVDASTDFSDTTSGSLNLLTPSSTTSRVRDFTSRIQERAGSNLQQNYLTPTPVVTPIRTPEETTPTPRKSEHLLKPPPKVHVDSSSHRSRGQHKRRDDLPQKKPSYLNLACSVNGYTNLTTYDSKLRQDINKSREASPIRPITYTYQYRSENNSLLVPVPVSANKLLVPTFGPHDTTDLTAKAPSKAHTDPSLATSTLHVYMAAENKQLKDDFLGPDMTTTSRPYISTESKNFATSMLHQKDELDNAKEITFKSSYSETNFRKTVTSNGKDSRFTSESYTISSNGMSKRVEITKENGEKLTSPMKSFIQQRVERLYGPGALAQGFFNQKRLKSQSEDDDSKVLTEKSLNCPEYSVRKLNKSFDDAYTSPNGNLDSSVSLPVLRHLRPEFRAQLPIHSPKKSVKAELSPQKLEEDIPRLDTKTELSEVVTNGVSVIKIDDSLNNDAVNGKEINGEAVKDAYYFLDLEKKETERLLALSASAEKELENLQVITTFTF
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF03359 GKAP
Interpro
IPR005026
SAPAP
ProteinModelPortal
Ontologies
GO
PANTHER
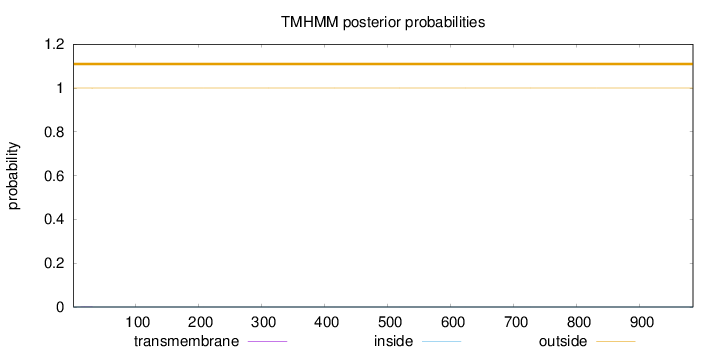
Topology
Length:
984
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00323
Exp number, first 60 AAs:
0.00323
Total prob of N-in:
0.00017
outside
1 - 984
Population Genetic Test Statistics
Pi
223.895156
Theta
199.440266
Tajima's D
0.930263
CLR
0.218336
CSRT
0.643317834108295
Interpretation
Uncertain
