Gene
KWMTBOMO00884
Pre Gene Modal
BGIBMGA009773
Annotation
PREDICTED:_KAT8_regulatory_NSL_complex_subunit_2_isoform_X2_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 4.717
Sequence
CDS
ATGACGTCACAACCTCGTAAATTAATTCACCTGCCCAAAGTCAGGATGCTCAGTAGTGGGGGACCAACAAAATCCGGTATCCGGATAACAAATGTGAGGACTATAAAGCCTCCAGACCCGGAACACGTGAAGAAACAGGAAGAGGAGAAACTGAGGGCTCAGGTCCAAAAGGAGATCGCCTCGCGGTGTCGCTCGTGCGCGTACAAGGGCTACGAATGCTTGCAGCCCGTGCTGTCTGGTCGGCAGTACTGCTACAGACATGTGCTCCGCGACCCGACCGCCCCCTACAGGCAGTGCGTGCACAACTATGCCAACGGGGAGAGGTGTACGTTGCCGGCACCCATGGACACTAATGATCCCCGGGACCCTGGGTTGTGTTTCGAGCACGCCCGCGCCGCCCTCGCCCTCCGCCAGCGCCAGGCCGCGCCCCCTCCGCCCCTCCCCACCACCGAGACGCTGCTCCACCAGCTCAGCCACTACATACAAGCAGAGAGGCCCCGCACGACGTCATGTGCGTCTTCAGTGTCCGTTGTAAGCGACCCATCGGAACCAGAACAGTCTTCTCCGACAGTACTAGATCCATTCAAACAAATAGACGCGAGGTCGGTGAATGCATCGTACTCAGCGTCCATACAAGAGTACTGCAGTGCCTCTGACAGTGACGGTGACAGTGTCAAGCTGGGGGAAAACGGGGAATGTAGCTCGGGGGACGATGAACTCGCACCTGTTGAAGAGCTACCATTATGGCGTGCCGGTGTGTACACCGCCGAAGAAACTGTAAGTGAAGCGAGAAATGCACTAAAACTATTGCAAATGGCGTACGTCAATCAAATGGGGCGTCTCCGGGTGCTGCTACAGATGTCCCGGGCTAAATACGTGAAGGCTCTCAAGGCTGAGAAGGAACACTACTGCAGTATAAACACGCAAACCCGCAACGGTCCGCCCACGGCTCGCGAGAGGAGGCAACTGCGGAAACTGAAGGCCTTTGCTTCATATCACAAGAAACACGGTGTGGATGCTGTGCTCGGCAGGAAGCTACATCATAAGAGAGCCAAAGTCAACGACCCACACCCATCCCGGCCGGTGCCGTCCACGTGTCGCTGCACGTTCTCCGAGGGCGGCGTGCGGTGCCCCGCGCAGGCGCTGCCGGCCGCCAAGCACTGCCTCAAGCATATACTGCACGACAGACATCAAGTAACCTCCCTCAATCCACTATATGAGTCGTCTATTATCAAAGGTGGCAATCTGTGCATTTGGACAAAAAAATGTTATTGA
Protein
MTSQPRKLIHLPKVRMLSSGGPTKSGIRITNVRTIKPPDPEHVKKQEEEKLRAQVQKEIASRCRSCAYKGYECLQPVLSGRQYCYRHVLRDPTAPYRQCVHNYANGERCTLPAPMDTNDPRDPGLCFEHARAALALRQRQAAPPPPLPTTETLLHQLSHYIQAERPRTTSCASSVSVVSDPSEPEQSSPTVLDPFKQIDARSVNASYSASIQEYCSASDSDGDSVKLGENGECSSGDDELAPVEELPLWRAGVYTAEETVSEARNALKLLQMAYVNQMGRLRVLLQMSRAKYVKALKAEKEHYCSINTQTRNGPPTARERRQLRKLKAFASYHKKHGVDAVLGRKLHHKRAKVNDPHPSRPVPSTCRCTFSEGGVRCPAQALPAAKHCLKHILHDRHQVTSLNPLYESSIIKGGNLCIWTKKCY
Summary
Uniprot
H9JJS2
A0A194PTG3
A0A194R577
A0A212ELB6
A0A1W4WMY1
A0A139WFM9
+ More
A0A139WFI1 E2C329 A0A310SK12 A0A0N0BBV6 A0A0L7RFG6 A0A154P1C8 A0A1B0GKN2 A0A0J7KD86 A0A195FKB1 K7ITK8 A0A1L8DKF7 A0A151I4S3 A0A158NU00 A0A151WMY8 A0A232F823 E9IQ47 A0A195C7C3 A0A1Y1N652 A7UUI1 A0A1Y1MZM0 A0A3L8DSJ4 A0A182PHR8 A0A1Y1N447 A0A1Y1N425 A0A1Y1MZN5 A0A1Y1N2J3 A0A1Y1MZM9 A0A182GKU1 A0A182V693 Q0IFG9 A0A1S4F8R4 A0A195DKL6 Q7Q521 A0A182I4Q6 A0A182KNA9 A0A0L0CNP7 A0A026X2V1 N6TJQ9 A0A1B0FCF6 A0A182Y1E1 B0XGU8 B0X0N1 A0A182WIE0 W5JC47 A0A182MFL4 A0A1Q3F3P5 A0A1Q3F3G8 A0A1Q3F370 A0A1A9VMM0 A0A1B0AIB7 T1P8F6 A0A1I8N400 A0A1B0AXV0 A0A182FFR1 A0A336MFJ7 A0A336KB73 A0A1A9Y9R7 A0A1A9WFD8 A0A084WSW7 A0A182J5J3 A0A182R2E7 A0A182QD86 A0A2P8XKY8 E0VP98 A0A182N7S3 A0A1I8PVR7 A0A034WCJ9 A0A0K8TLF7 A0A1B6CND5 A0A1B6DHY2 A0A1B6C089 T1GRI2 A0A034W1C3 A0A2M4ABT2 A0A2M4ABS6 A0A034W3Y4 A0A0A1XQW7 A0A0K8TXI1 A0A0A1X894 A0A0A1WRZ4 A0A0K8VZA6 W8C5A2 W8CBA5 W8BJS2 E2B259 A0A0Q9WC04
A0A139WFI1 E2C329 A0A310SK12 A0A0N0BBV6 A0A0L7RFG6 A0A154P1C8 A0A1B0GKN2 A0A0J7KD86 A0A195FKB1 K7ITK8 A0A1L8DKF7 A0A151I4S3 A0A158NU00 A0A151WMY8 A0A232F823 E9IQ47 A0A195C7C3 A0A1Y1N652 A7UUI1 A0A1Y1MZM0 A0A3L8DSJ4 A0A182PHR8 A0A1Y1N447 A0A1Y1N425 A0A1Y1MZN5 A0A1Y1N2J3 A0A1Y1MZM9 A0A182GKU1 A0A182V693 Q0IFG9 A0A1S4F8R4 A0A195DKL6 Q7Q521 A0A182I4Q6 A0A182KNA9 A0A0L0CNP7 A0A026X2V1 N6TJQ9 A0A1B0FCF6 A0A182Y1E1 B0XGU8 B0X0N1 A0A182WIE0 W5JC47 A0A182MFL4 A0A1Q3F3P5 A0A1Q3F3G8 A0A1Q3F370 A0A1A9VMM0 A0A1B0AIB7 T1P8F6 A0A1I8N400 A0A1B0AXV0 A0A182FFR1 A0A336MFJ7 A0A336KB73 A0A1A9Y9R7 A0A1A9WFD8 A0A084WSW7 A0A182J5J3 A0A182R2E7 A0A182QD86 A0A2P8XKY8 E0VP98 A0A182N7S3 A0A1I8PVR7 A0A034WCJ9 A0A0K8TLF7 A0A1B6CND5 A0A1B6DHY2 A0A1B6C089 T1GRI2 A0A034W1C3 A0A2M4ABT2 A0A2M4ABS6 A0A034W3Y4 A0A0A1XQW7 A0A0K8TXI1 A0A0A1X894 A0A0A1WRZ4 A0A0K8VZA6 W8C5A2 W8CBA5 W8BJS2 E2B259 A0A0Q9WC04
Pubmed
19121390
26354079
22118469
18362917
19820115
20798317
+ More
20075255 21347285 28648823 21282665 28004739 12364791 30249741 26483478 17510324 14747013 17210077 20966253 26108605 24508170 23537049 25244985 20920257 23761445 25315136 24438588 29403074 20566863 25348373 26369729 25830018 24495485 17994087
20075255 21347285 28648823 21282665 28004739 12364791 30249741 26483478 17510324 14747013 17210077 20966253 26108605 24508170 23537049 25244985 20920257 23761445 25315136 24438588 29403074 20566863 25348373 26369729 25830018 24495485 17994087
EMBL
BABH01017691
BABH01017692
BABH01017693
KQ459593
KPI96597.1
KQ460890
+ More
KPJ11016.1 AGBW02014100 OWR42261.1 KQ971352 KYB26724.1 KYB26723.1 GL452274 EFN77645.1 KQ760512 OAD60052.1 KQ435970 KOX67798.1 KQ414608 KOC69466.1 KQ434796 KZC05643.1 AJWK01030744 LBMM01009420 KMQ88149.1 KQ981522 KYN40697.1 GFDF01007227 JAV06857.1 KQ976445 KYM86080.1 ADTU01026230 KQ982926 KYQ49226.1 NNAY01000761 OXU26640.1 GL764659 EFZ17328.1 KQ978143 KYM96774.1 GEZM01016859 GEZM01016857 GEZM01016854 JAV91107.1 AAAB01008960 EDO63959.1 GEZM01016861 JAV91099.1 QOIP01000004 RLU23381.1 GEZM01016860 GEZM01016856 GEZM01016852 JAV91116.1 GEZM01016862 GEZM01016853 JAV91096.1 GEZM01016851 JAV91119.1 GEZM01016855 JAV91110.1 GEZM01016858 JAV91104.1 JXUM01014101 KQ560394 KXJ82637.1 CH477317 EAT43746.1 KQ980794 KYN13034.1 EAA11850.4 APCN01002346 JRES01000141 KNC33876.1 KK107046 EZA61714.1 APGK01034947 KB740923 KB632430 ENN78098.1 ERL95492.1 CCAG010010775 DS233065 EDS27835.1 DS232242 EDS38273.1 ADMH02001932 ETN60455.1 AXCM01001108 GFDL01012882 JAV22163.1 GFDL01012976 JAV22069.1 GFDL01013076 JAV21969.1 KA645006 AFP59635.1 JXJN01005427 UFQT01001144 SSX29134.1 UFQS01000258 UFQT01000258 SSX02126.1 SSX22503.1 ATLV01026739 KE525418 KFB53311.1 AXCN02000584 PYGN01001820 PSN32668.1 DS235361 EEB15203.1 GAKP01007434 JAC51518.1 GDAI01002416 JAI15187.1 GEDC01022356 JAS14942.1 GEDC01012063 JAS25235.1 GEDC01030514 JAS06784.1 CAQQ02089189 GAKP01010468 JAC48484.1 GGFK01004923 MBW38244.1 GGFK01004922 MBW38243.1 GAKP01010469 JAC48483.1 GBXI01010223 GBXI01000583 JAD04069.1 JAD13709.1 GDHF01033554 JAI18760.1 GBXI01006985 JAD07307.1 GBXI01012488 JAD01804.1 GDHF01008394 JAI43920.1 GAMC01005004 JAC01552.1 GAMC01005006 JAC01550.1 GAMC01005005 JAC01551.1 GL445059 EFN60234.1 CH940656 KRF78350.1
KPJ11016.1 AGBW02014100 OWR42261.1 KQ971352 KYB26724.1 KYB26723.1 GL452274 EFN77645.1 KQ760512 OAD60052.1 KQ435970 KOX67798.1 KQ414608 KOC69466.1 KQ434796 KZC05643.1 AJWK01030744 LBMM01009420 KMQ88149.1 KQ981522 KYN40697.1 GFDF01007227 JAV06857.1 KQ976445 KYM86080.1 ADTU01026230 KQ982926 KYQ49226.1 NNAY01000761 OXU26640.1 GL764659 EFZ17328.1 KQ978143 KYM96774.1 GEZM01016859 GEZM01016857 GEZM01016854 JAV91107.1 AAAB01008960 EDO63959.1 GEZM01016861 JAV91099.1 QOIP01000004 RLU23381.1 GEZM01016860 GEZM01016856 GEZM01016852 JAV91116.1 GEZM01016862 GEZM01016853 JAV91096.1 GEZM01016851 JAV91119.1 GEZM01016855 JAV91110.1 GEZM01016858 JAV91104.1 JXUM01014101 KQ560394 KXJ82637.1 CH477317 EAT43746.1 KQ980794 KYN13034.1 EAA11850.4 APCN01002346 JRES01000141 KNC33876.1 KK107046 EZA61714.1 APGK01034947 KB740923 KB632430 ENN78098.1 ERL95492.1 CCAG010010775 DS233065 EDS27835.1 DS232242 EDS38273.1 ADMH02001932 ETN60455.1 AXCM01001108 GFDL01012882 JAV22163.1 GFDL01012976 JAV22069.1 GFDL01013076 JAV21969.1 KA645006 AFP59635.1 JXJN01005427 UFQT01001144 SSX29134.1 UFQS01000258 UFQT01000258 SSX02126.1 SSX22503.1 ATLV01026739 KE525418 KFB53311.1 AXCN02000584 PYGN01001820 PSN32668.1 DS235361 EEB15203.1 GAKP01007434 JAC51518.1 GDAI01002416 JAI15187.1 GEDC01022356 JAS14942.1 GEDC01012063 JAS25235.1 GEDC01030514 JAS06784.1 CAQQ02089189 GAKP01010468 JAC48484.1 GGFK01004923 MBW38244.1 GGFK01004922 MBW38243.1 GAKP01010469 JAC48483.1 GBXI01010223 GBXI01000583 JAD04069.1 JAD13709.1 GDHF01033554 JAI18760.1 GBXI01006985 JAD07307.1 GBXI01012488 JAD01804.1 GDHF01008394 JAI43920.1 GAMC01005004 JAC01552.1 GAMC01005006 JAC01550.1 GAMC01005005 JAC01551.1 GL445059 EFN60234.1 CH940656 KRF78350.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000192223
UP000007266
+ More
UP000008237 UP000053105 UP000053825 UP000076502 UP000092461 UP000036403 UP000078541 UP000002358 UP000078540 UP000005205 UP000075809 UP000215335 UP000078542 UP000007062 UP000279307 UP000075885 UP000069940 UP000249989 UP000075903 UP000008820 UP000078492 UP000075840 UP000075882 UP000037069 UP000053097 UP000019118 UP000030742 UP000092444 UP000076408 UP000002320 UP000075920 UP000000673 UP000075883 UP000078200 UP000092445 UP000095301 UP000092460 UP000069272 UP000092443 UP000091820 UP000030765 UP000075880 UP000075900 UP000075886 UP000245037 UP000009046 UP000075884 UP000095300 UP000015102 UP000000311 UP000008792
UP000008237 UP000053105 UP000053825 UP000076502 UP000092461 UP000036403 UP000078541 UP000002358 UP000078540 UP000005205 UP000075809 UP000215335 UP000078542 UP000007062 UP000279307 UP000075885 UP000069940 UP000249989 UP000075903 UP000008820 UP000078492 UP000075840 UP000075882 UP000037069 UP000053097 UP000019118 UP000030742 UP000092444 UP000076408 UP000002320 UP000075920 UP000000673 UP000075883 UP000078200 UP000092445 UP000095301 UP000092460 UP000069272 UP000092443 UP000091820 UP000030765 UP000075880 UP000075900 UP000075886 UP000245037 UP000009046 UP000075884 UP000095300 UP000015102 UP000000311 UP000008792
PRIDE
Pfam
PF13891 zf-C3Hc3H
ProteinModelPortal
H9JJS2
A0A194PTG3
A0A194R577
A0A212ELB6
A0A1W4WMY1
A0A139WFM9
+ More
A0A139WFI1 E2C329 A0A310SK12 A0A0N0BBV6 A0A0L7RFG6 A0A154P1C8 A0A1B0GKN2 A0A0J7KD86 A0A195FKB1 K7ITK8 A0A1L8DKF7 A0A151I4S3 A0A158NU00 A0A151WMY8 A0A232F823 E9IQ47 A0A195C7C3 A0A1Y1N652 A7UUI1 A0A1Y1MZM0 A0A3L8DSJ4 A0A182PHR8 A0A1Y1N447 A0A1Y1N425 A0A1Y1MZN5 A0A1Y1N2J3 A0A1Y1MZM9 A0A182GKU1 A0A182V693 Q0IFG9 A0A1S4F8R4 A0A195DKL6 Q7Q521 A0A182I4Q6 A0A182KNA9 A0A0L0CNP7 A0A026X2V1 N6TJQ9 A0A1B0FCF6 A0A182Y1E1 B0XGU8 B0X0N1 A0A182WIE0 W5JC47 A0A182MFL4 A0A1Q3F3P5 A0A1Q3F3G8 A0A1Q3F370 A0A1A9VMM0 A0A1B0AIB7 T1P8F6 A0A1I8N400 A0A1B0AXV0 A0A182FFR1 A0A336MFJ7 A0A336KB73 A0A1A9Y9R7 A0A1A9WFD8 A0A084WSW7 A0A182J5J3 A0A182R2E7 A0A182QD86 A0A2P8XKY8 E0VP98 A0A182N7S3 A0A1I8PVR7 A0A034WCJ9 A0A0K8TLF7 A0A1B6CND5 A0A1B6DHY2 A0A1B6C089 T1GRI2 A0A034W1C3 A0A2M4ABT2 A0A2M4ABS6 A0A034W3Y4 A0A0A1XQW7 A0A0K8TXI1 A0A0A1X894 A0A0A1WRZ4 A0A0K8VZA6 W8C5A2 W8CBA5 W8BJS2 E2B259 A0A0Q9WC04
A0A139WFI1 E2C329 A0A310SK12 A0A0N0BBV6 A0A0L7RFG6 A0A154P1C8 A0A1B0GKN2 A0A0J7KD86 A0A195FKB1 K7ITK8 A0A1L8DKF7 A0A151I4S3 A0A158NU00 A0A151WMY8 A0A232F823 E9IQ47 A0A195C7C3 A0A1Y1N652 A7UUI1 A0A1Y1MZM0 A0A3L8DSJ4 A0A182PHR8 A0A1Y1N447 A0A1Y1N425 A0A1Y1MZN5 A0A1Y1N2J3 A0A1Y1MZM9 A0A182GKU1 A0A182V693 Q0IFG9 A0A1S4F8R4 A0A195DKL6 Q7Q521 A0A182I4Q6 A0A182KNA9 A0A0L0CNP7 A0A026X2V1 N6TJQ9 A0A1B0FCF6 A0A182Y1E1 B0XGU8 B0X0N1 A0A182WIE0 W5JC47 A0A182MFL4 A0A1Q3F3P5 A0A1Q3F3G8 A0A1Q3F370 A0A1A9VMM0 A0A1B0AIB7 T1P8F6 A0A1I8N400 A0A1B0AXV0 A0A182FFR1 A0A336MFJ7 A0A336KB73 A0A1A9Y9R7 A0A1A9WFD8 A0A084WSW7 A0A182J5J3 A0A182R2E7 A0A182QD86 A0A2P8XKY8 E0VP98 A0A182N7S3 A0A1I8PVR7 A0A034WCJ9 A0A0K8TLF7 A0A1B6CND5 A0A1B6DHY2 A0A1B6C089 T1GRI2 A0A034W1C3 A0A2M4ABT2 A0A2M4ABS6 A0A034W3Y4 A0A0A1XQW7 A0A0K8TXI1 A0A0A1X894 A0A0A1WRZ4 A0A0K8VZA6 W8C5A2 W8CBA5 W8BJS2 E2B259 A0A0Q9WC04
Ontologies
PANTHER
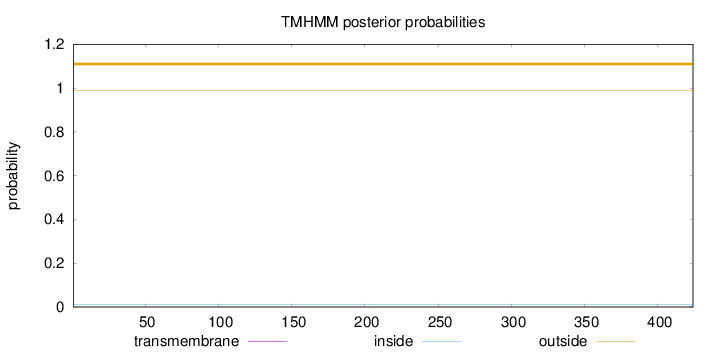
Topology
Length:
424
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00138
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01217
outside
1 - 424
Population Genetic Test Statistics
Pi
212.529607
Theta
172.317586
Tajima's D
0.860495
CLR
0.742418
CSRT
0.625718714064297
Interpretation
Uncertain
