Gene
KWMTBOMO00881 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009770
Annotation
PREDICTED:_ced-6_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.251
Sequence
CDS
ATGTCGACCCTGCTCTTCTGGCAGGGTAAGGGCAAAGGGAACGGGGCACCGAACGGCCGCAACTGGATCCACGCTCCGGACTCGCTCGTCAAGGGCCACGTCGCCTATCTCGTTAAGTTCCTCGGCTGCACCCAAGTAGACCAGCCGAAAGGCATCGAGGTGGTCAAAGATGCAATCAAGAAACTACAGTTCACACAGCAACTGAAGAAGTCCGAAGCGAAAGATGGCGCGAAATGCAAGAAAGTCGAAATAACGATATCTGTGGACGGTGTTGCCATACAGGAACCACGTTCGAACAATATAATGTACCAGTTCCCTCTCCACCGGATATCGTACTGCGCGGACGATAAAGGCGCCAAGAAATATTTCTCGTTCATCGCGAAAGGTGGCAGCACCGTTAACGGGGTCAACGGCCACGACGCTGGCAACACCGAGAGGCACGAGTGCTTCGTGTTCATTTCCACTAAACTCGCGTCCGAGATCACGCTCACCATCGGACAGGCCTTCGATCTGGCCTACAGGAGATTCTTAAACGACAACGGCAAATACATCGAAATGCAGAAGCTTTCGTTGCAAAACAAAAAGCTTGAACTGCAAATGAAAGCGTACAGGACTAGACTTCAGGAGCTGTCGAAGGTCACGTATAAAGACGACCTCTCCGCCTACCTGGCCAGGAACAGTCTCTCCGACATAACAGAGTTCAAGGTACCCCAGGAGTTGGAGAAGCAGCTCGCCTCGCTGACCTTAGCGAACGGCTTCAGCGCCATGAACGGGAACAAGACCCCTGAAATCGGCACAGACTCCACATCAAACAACTCAACGGACACGTTCAATTCAAGCGACAACAATCTGCTTCTCCCCACCCCGCCGCCGACACATAATGGGAAGATGAGCGCCGGTAAGGGACTCCTCGATCTGCCAAAGAATCCCGTCGTAGGTCCGAGACTTGAGGAGCTAATACTCAACTCAGATTCGGACAGCGATTTTGATCCTCGCGCCCACGAATCGGACCCCGTCCTCCGCTACCCAGGTCTGACCATGGACAGCACGGTGTCACCGGCGCCTCCGTTACTGGCGCCGCCGCCTAAGGCGTCGAAACGTCAAACGCCGCCGCACAACCCGGCGACGCCGCCGCACCAGCCGACGACGCCGCGGCGAGCGCCCGCCGCGAACCCGAGCGACCTGTTCGGCTCGGCGCCGTTCGTGCACAGCTCTACGCCCGCGCAAGCTCCGTTCGCGCCGACGCCCAACTACAACATAAGCGACTTCAACCTCCAAACATCGGTGTTCACGGCGAACCTCACCGGCTACGATCCGTTCGAAGCTCAGAAGACGGGCGCGAACATATTCGGGAACGATTACGCGAACGCGTTCGCCGGCTTCGGGAATCTGAGCGACAAACAGACGGTCGAATTGAACAGCAGCTTCAACGGGTTCCTAGATAAGACCATATCGGAGATGAAGGACGGATTCACGAGGGGCATAACGTTCGGAAACGACGATTTCTCAATAGACAATCTCGATCCTTTAAAGAAAAATTAA
Protein
MSTLLFWQGKGKGNGAPNGRNWIHAPDSLVKGHVAYLVKFLGCTQVDQPKGIEVVKDAIKKLQFTQQLKKSEAKDGAKCKKVEITISVDGVAIQEPRSNNIMYQFPLHRISYCADDKGAKKYFSFIAKGGSTVNGVNGHDAGNTERHECFVFISTKLASEITLTIGQAFDLAYRRFLNDNGKYIEMQKLSLQNKKLELQMKAYRTRLQELSKVTYKDDLSAYLARNSLSDITEFKVPQELEKQLASLTLANGFSAMNGNKTPEIGTDSTSNNSTDTFNSSDNNLLLPTPPPTHNGKMSAGKGLLDLPKNPVVGPRLEELILNSDSDSDFDPRAHESDPVLRYPGLTMDSTVSPAPPLLAPPPKASKRQTPPHNPATPPHQPTTPRRAPAANPSDLFGSAPFVHSSTPAQAPFAPTPNYNISDFNLQTSVFTANLTGYDPFEAQKTGANIFGNDYANAFAGFGNLSDKQTVELNSSFNGFLDKTISEMKDGFTRGITFGNDDFSIDNLDPLKKN
Summary
Uniprot
D4QF47
H9JJR9
A0A2H1W0A5
A0A2A4IWS5
S4PTE1
A0A212F6H3
+ More
A0A194QZY4 A0A336LRE2 A0A336LM05 A0A1J1HHY9 U5EZS3 A0A084WKQ0 Q7QIK3 A0A2M3YZC8 W5JFH0 A0A1Y1LEH8 A0A2M4CKK0 A0A2M4A0X0 A0A2M4CKQ6 A0A2M3YZ35 D6WT62 A0A182KAB5 A0A182PIK9 A0A182JI39 A0A2M4CIM8 A0A182FEH4 A0A2M3YZ83 A0A2M4A0V9 A0NBM1 A0A2M4A0W3 A0A182GMF1 A0A182TI04 A0A182I6P3 A0A182WWG3 A0A182GTK5 A0A182US69 A0A182L3P9 T1E7P0 A0A182R0U8 A0A182Y7Z5 A0A2A3E8B8 E2AUL6 A0A182MT38 A0A026W3P6 A0A182N9D1 A0A182WBG4 A0A182RJI9 A0A0L7RCD6 A0A1B6CS71 A0A0T6BCG4 A0A310SJA6 V9IC64 A0A0C9QKM5 A0A087ZS23 A0A0M9A8J0 E2BQ59 W8C471 A0A034W0N5 B4MFH3 J9KYP9 A0A0A1XK59
A0A194QZY4 A0A336LRE2 A0A336LM05 A0A1J1HHY9 U5EZS3 A0A084WKQ0 Q7QIK3 A0A2M3YZC8 W5JFH0 A0A1Y1LEH8 A0A2M4CKK0 A0A2M4A0X0 A0A2M4CKQ6 A0A2M3YZ35 D6WT62 A0A182KAB5 A0A182PIK9 A0A182JI39 A0A2M4CIM8 A0A182FEH4 A0A2M3YZ83 A0A2M4A0V9 A0NBM1 A0A2M4A0W3 A0A182GMF1 A0A182TI04 A0A182I6P3 A0A182WWG3 A0A182GTK5 A0A182US69 A0A182L3P9 T1E7P0 A0A182R0U8 A0A182Y7Z5 A0A2A3E8B8 E2AUL6 A0A182MT38 A0A026W3P6 A0A182N9D1 A0A182WBG4 A0A182RJI9 A0A0L7RCD6 A0A1B6CS71 A0A0T6BCG4 A0A310SJA6 V9IC64 A0A0C9QKM5 A0A087ZS23 A0A0M9A8J0 E2BQ59 W8C471 A0A034W0N5 B4MFH3 J9KYP9 A0A0A1XK59
Pubmed
EMBL
AB531437
BAJ04619.1
BABH01017682
BABH01017683
BABH01017684
ODYU01005512
+ More
SOQ46433.1 NWSH01005373 PCG64265.1 GAIX01012198 JAA80362.1 AGBW02010043 OWR49309.1 KQ460890 KPJ11012.1 UFQT01000063 SSX19273.1 SSX19274.1 CVRI01000003 CRK87028.1 GANO01000253 JAB59618.1 ATLV01024132 KE525349 KFB50794.1 AAAB01008807 EAA04093.4 GGFM01000817 MBW21568.1 ADMH02001370 ETN62821.1 GEZM01058010 GEZM01058009 JAV72024.1 GGFL01001704 MBW65882.1 GGFK01001102 MBW34423.1 GGFL01001705 MBW65883.1 GGFM01000765 MBW21516.1 KQ971354 EFA07168.1 GGFL01001034 MBW65212.1 GGFM01000826 MBW21577.1 GGFK01001078 MBW34399.1 EAU77688.2 GGFK01001133 MBW34454.1 JXUM01074562 JXUM01074563 KQ562837 KXJ75039.1 APCN01003688 JXUM01087381 KQ563629 KXJ73578.1 GAMD01002877 JAA98713.1 AXCN02000560 KZ288340 PBC27744.1 GL442829 EFN62863.1 AXCM01010355 AXCM01010356 KK107447 QOIP01000001 EZA50710.1 RLU26345.1 KQ414615 KOC68657.1 GEDC01021014 JAS16284.1 LJIG01001916 KRT85024.1 KQ764883 OAD54266.1 JR038422 AEY58247.1 GBYB01015338 JAG85105.1 KQ435711 KOX79407.1 GL449698 EFN82178.1 GAMC01005717 GAMC01005716 GAMC01005715 GAMC01005714 JAC00842.1 GAKP01009793 GAKP01009792 GAKP01009791 JAC49161.1 CH940667 EDW57144.1 KRF77625.1 KRF77626.1 ABLF02031199 ABLF02031200 GBXI01016705 GBXI01012555 GBXI01003032 JAC97586.1 JAD01737.1 JAD11260.1
SOQ46433.1 NWSH01005373 PCG64265.1 GAIX01012198 JAA80362.1 AGBW02010043 OWR49309.1 KQ460890 KPJ11012.1 UFQT01000063 SSX19273.1 SSX19274.1 CVRI01000003 CRK87028.1 GANO01000253 JAB59618.1 ATLV01024132 KE525349 KFB50794.1 AAAB01008807 EAA04093.4 GGFM01000817 MBW21568.1 ADMH02001370 ETN62821.1 GEZM01058010 GEZM01058009 JAV72024.1 GGFL01001704 MBW65882.1 GGFK01001102 MBW34423.1 GGFL01001705 MBW65883.1 GGFM01000765 MBW21516.1 KQ971354 EFA07168.1 GGFL01001034 MBW65212.1 GGFM01000826 MBW21577.1 GGFK01001078 MBW34399.1 EAU77688.2 GGFK01001133 MBW34454.1 JXUM01074562 JXUM01074563 KQ562837 KXJ75039.1 APCN01003688 JXUM01087381 KQ563629 KXJ73578.1 GAMD01002877 JAA98713.1 AXCN02000560 KZ288340 PBC27744.1 GL442829 EFN62863.1 AXCM01010355 AXCM01010356 KK107447 QOIP01000001 EZA50710.1 RLU26345.1 KQ414615 KOC68657.1 GEDC01021014 JAS16284.1 LJIG01001916 KRT85024.1 KQ764883 OAD54266.1 JR038422 AEY58247.1 GBYB01015338 JAG85105.1 KQ435711 KOX79407.1 GL449698 EFN82178.1 GAMC01005717 GAMC01005716 GAMC01005715 GAMC01005714 JAC00842.1 GAKP01009793 GAKP01009792 GAKP01009791 JAC49161.1 CH940667 EDW57144.1 KRF77625.1 KRF77626.1 ABLF02031199 ABLF02031200 GBXI01016705 GBXI01012555 GBXI01003032 JAC97586.1 JAD01737.1 JAD11260.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000183832
UP000030765
+ More
UP000007062 UP000000673 UP000007266 UP000075881 UP000075885 UP000075880 UP000069272 UP000069940 UP000249989 UP000075902 UP000075840 UP000076407 UP000075903 UP000075882 UP000075886 UP000076408 UP000242457 UP000000311 UP000075883 UP000053097 UP000279307 UP000075884 UP000075920 UP000075900 UP000053825 UP000005203 UP000053105 UP000008237 UP000008792 UP000007819
UP000007062 UP000000673 UP000007266 UP000075881 UP000075885 UP000075880 UP000069272 UP000069940 UP000249989 UP000075902 UP000075840 UP000076407 UP000075903 UP000075882 UP000075886 UP000076408 UP000242457 UP000000311 UP000075883 UP000053097 UP000279307 UP000075884 UP000075920 UP000075900 UP000053825 UP000005203 UP000053105 UP000008237 UP000008792 UP000007819
Pfam
PF00640 PID
Gene 3D
ProteinModelPortal
D4QF47
H9JJR9
A0A2H1W0A5
A0A2A4IWS5
S4PTE1
A0A212F6H3
+ More
A0A194QZY4 A0A336LRE2 A0A336LM05 A0A1J1HHY9 U5EZS3 A0A084WKQ0 Q7QIK3 A0A2M3YZC8 W5JFH0 A0A1Y1LEH8 A0A2M4CKK0 A0A2M4A0X0 A0A2M4CKQ6 A0A2M3YZ35 D6WT62 A0A182KAB5 A0A182PIK9 A0A182JI39 A0A2M4CIM8 A0A182FEH4 A0A2M3YZ83 A0A2M4A0V9 A0NBM1 A0A2M4A0W3 A0A182GMF1 A0A182TI04 A0A182I6P3 A0A182WWG3 A0A182GTK5 A0A182US69 A0A182L3P9 T1E7P0 A0A182R0U8 A0A182Y7Z5 A0A2A3E8B8 E2AUL6 A0A182MT38 A0A026W3P6 A0A182N9D1 A0A182WBG4 A0A182RJI9 A0A0L7RCD6 A0A1B6CS71 A0A0T6BCG4 A0A310SJA6 V9IC64 A0A0C9QKM5 A0A087ZS23 A0A0M9A8J0 E2BQ59 W8C471 A0A034W0N5 B4MFH3 J9KYP9 A0A0A1XK59
A0A194QZY4 A0A336LRE2 A0A336LM05 A0A1J1HHY9 U5EZS3 A0A084WKQ0 Q7QIK3 A0A2M3YZC8 W5JFH0 A0A1Y1LEH8 A0A2M4CKK0 A0A2M4A0X0 A0A2M4CKQ6 A0A2M3YZ35 D6WT62 A0A182KAB5 A0A182PIK9 A0A182JI39 A0A2M4CIM8 A0A182FEH4 A0A2M3YZ83 A0A2M4A0V9 A0NBM1 A0A2M4A0W3 A0A182GMF1 A0A182TI04 A0A182I6P3 A0A182WWG3 A0A182GTK5 A0A182US69 A0A182L3P9 T1E7P0 A0A182R0U8 A0A182Y7Z5 A0A2A3E8B8 E2AUL6 A0A182MT38 A0A026W3P6 A0A182N9D1 A0A182WBG4 A0A182RJI9 A0A0L7RCD6 A0A1B6CS71 A0A0T6BCG4 A0A310SJA6 V9IC64 A0A0C9QKM5 A0A087ZS23 A0A0M9A8J0 E2BQ59 W8C471 A0A034W0N5 B4MFH3 J9KYP9 A0A0A1XK59
PDB
3SO6
E-value=7.52055e-13,
Score=180
Ontologies
KEGG
GO
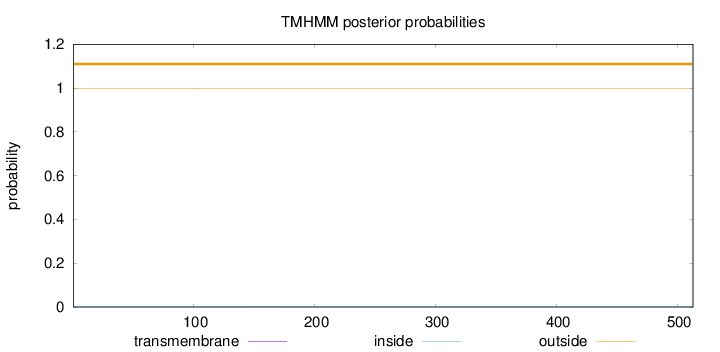
Topology
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00685999999999999
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00114
outside
1 - 513
Population Genetic Test Statistics
Pi
237.439112
Theta
164.89984
Tajima's D
0.512792
CLR
1.964358
CSRT
0.520273986300685
Interpretation
Uncertain
