Gene
KWMTBOMO00872 Validated by peptides from experiments
Annotation
chymotrypsin_inhibitor_SCI-III_precursor_[Bombyx_mori]
Full name
Chymotrypsin inhibitor SCI-III
+ More
Chymotrypsin inhibitor SCI-I
Chymotrypsin inhibitor SCI-II
Tissue factor pathway inhibitor
Kunitz-type serine protease inhibitor conotoxin Cal9.1a
Chymotrypsin inhibitor SCI-I
Chymotrypsin inhibitor SCI-II
Tissue factor pathway inhibitor
Kunitz-type serine protease inhibitor conotoxin Cal9.1a
Location in the cell
Extracellular Reliability : 3.865
Sequence
CDS
ATGTTGAGATCAGTGCTTTTTCTATTCGCGGGCGTTTTCTTCGCTGCGTGCTTGACATCGGTCATGAGTGATGAGCCAACGACGGACTTGCCGATATGCGAGCAGGCTTTTGGCGATGCGGGTCTTTGTTTCGGCTATATGAAACTATACTCGTACAACCAGGAGACGAAGAACTGTGAAGAATTCATTTACGGAGGCTGCCAAGGGAATGATAATCGTTTCAGCACATTGGCCGAATGCGAACAGAAATGCATAAACTAA
Protein
MLRSVLFLFAGVFFAACLTSVMSDEPTTDLPICEQAFGDAGLCFGYMKLYSYNQETKNCEEFIYGGCQGNDNRFSTLAECEQKCIN
Summary
Similarity
Belongs to the venom Kunitz-type family.
Keywords
Complete proteome
Direct protein sequencing
Disulfide bond
Protease inhibitor
Reference proteome
Serine protease inhibitor
Secreted
Signal
Toxin
Feature
chain Chymotrypsin inhibitor SCI-III
peptide Kunitz-type serine protease inhibitor conotoxin Cal9.1a
peptide Kunitz-type serine protease inhibitor conotoxin Cal9.1a
Uniprot
Pubmed
EMBL
AY167664
AAO17293.1
AF529176
AAN76278.1
AB062102
BAB83366.1
+ More
AF361484 AAK52496.1 AY839231 AY839232 AAW30166.1 AAW30167.2 KL367498 KFD69065.1 KN538402 KHJ42567.1 CH954177 EDV57666.1 CH963857 EDW76463.1 CH379060 EAL33748.2 CH479180 EDW28961.1 KFD69068.1 GU306152 GGLE01004376 MBY08502.1 AY437082 AAR97367.1 FR734169 CBY65969.1 GBBR01000066 JAC58996.1 PZQS01000008 PVD26538.1 GL446901 EFN87117.1 EDW76466.2 CH933807 EDW12596.1 AMQN01008243 KB302535 ELU04243.1 JQ730814 AFN01659.1 GAMC01004066 JAC02490.1 CH916372 EDV99167.1 LIAE01005567 PAV93241.1
AF361484 AAK52496.1 AY839231 AY839232 AAW30166.1 AAW30167.2 KL367498 KFD69065.1 KN538402 KHJ42567.1 CH954177 EDV57666.1 CH963857 EDW76463.1 CH379060 EAL33748.2 CH479180 EDW28961.1 KFD69068.1 GU306152 GGLE01004376 MBY08502.1 AY437082 AAR97367.1 FR734169 CBY65969.1 GBBR01000066 JAC58996.1 PZQS01000008 PVD26538.1 GL446901 EFN87117.1 EDW76466.2 CH933807 EDW12596.1 AMQN01008243 KB302535 ELU04243.1 JQ730814 AFN01659.1 GAMC01004066 JAC02490.1 CH916372 EDV99167.1 LIAE01005567 PAV93241.1
Proteomes
Pfam
Interpro
IPR020901
Prtase_inh_Kunz-CS
+ More
IPR036880 Kunitz_BPTI_sf
IPR002223 Kunitz_BPTI
IPR008296 TFPI-like
IPR002557 Chitin-bd_dom
IPR000980 SH2
IPR036860 SH2_dom_sf
IPR036383 TSP1_rpt_sf
IPR013106 Ig_V-set
IPR000884 TSP1_rpt
IPR036179 Ig-like_dom_sf
IPR010909 PLAC
IPR003598 Ig_sub2
IPR010294 ADAM_spacer1
IPR013783 Ig-like_fold
IPR000020 Anaphylatoxin/fibulin
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR008197 WAP_dom
IPR013273 ADAMTS/ADAMTS-like
IPR036645 Elafin-like_sf
IPR012674 Calycin
IPR036880 Kunitz_BPTI_sf
IPR002223 Kunitz_BPTI
IPR008296 TFPI-like
IPR002557 Chitin-bd_dom
IPR000980 SH2
IPR036860 SH2_dom_sf
IPR036383 TSP1_rpt_sf
IPR013106 Ig_V-set
IPR000884 TSP1_rpt
IPR036179 Ig-like_dom_sf
IPR010909 PLAC
IPR003598 Ig_sub2
IPR010294 ADAM_spacer1
IPR013783 Ig-like_fold
IPR000020 Anaphylatoxin/fibulin
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR008197 WAP_dom
IPR013273 ADAMTS/ADAMTS-like
IPR036645 Elafin-like_sf
IPR012674 Calycin
SUPFAM
CDD
ProteinModelPortal
PDB
5NMV
E-value=6.6398e-08,
Score=129
Ontologies
GO
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 23,
Likelihood: 0.999176
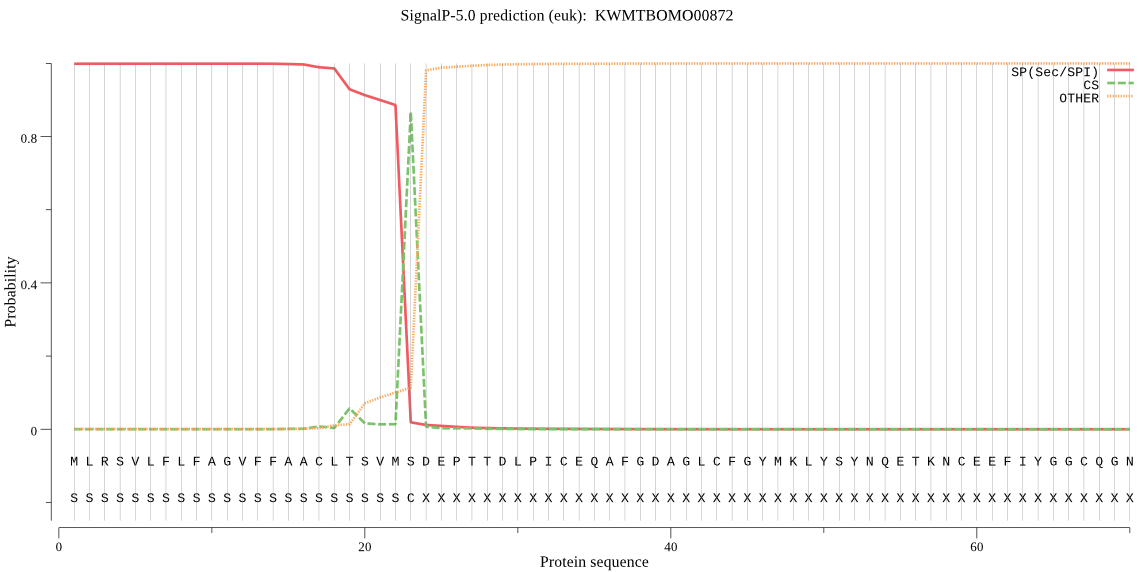
Length:
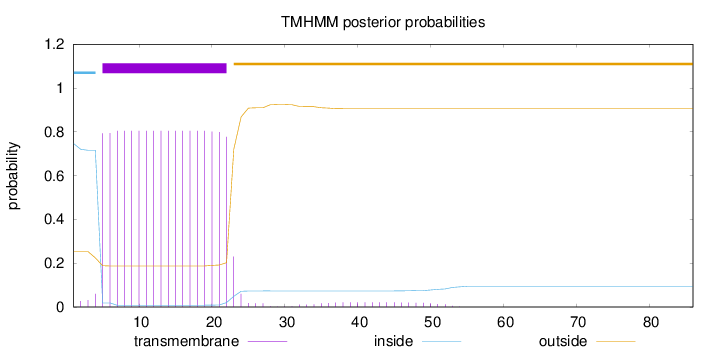
86
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.28214
Exp number, first 60 AAs:
15.28214
Total prob of N-in:
0.74691
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 22
outside
23 - 86
Population Genetic Test Statistics
Pi
234.843748
Theta
158.167737
Tajima's D
1.409583
CLR
0.027725
CSRT
0.770261486925654
Interpretation
Uncertain
