Gene
KWMTBOMO00860 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009760
Annotation
coatomer_protein_complex_subunit_zeta_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.317 Golgi Reliability : 1.849
Sequence
CDS
ATGGAAGGTTCTCTGTTCGAACCCACATTGTACATTGTCAAGGGGATGTGCATTTTGGACTATGAAGGAAACAGAATTTTGGCCAAATATTACGATAAAGACGTCCTACCCACAACTAAGGAGCAGAAGGCATTTGAGAAGAACTTGTTCAATAAGACTCACAGAGCAAATGCAGAAATCATAATGCTGGATGGTCTTACTTGTGTTTATAAAAGTAACGTAGATTTATTCTTCTATGTTATGGGAAGCTCCCATGAAAATGAGTTGATCCTTCAATCAGTATTGAATGCACTCTACGAGTCTGTTAGTTTGCTGCTAAGAAGGAACATGGAGCGGAGGGTCCTCATGGAGAACTTGGATGCTGTTATGCTTGCCTTCGATGAGATCTGCGACGGCGGTGTGATATTGGATGCTGATCCCACGTCGATCGTAAGCCGGGCAGCGCTGCGCACCGAGGACGTGCCCCTGGGAGAGCAAACAGTGGCCCAGGTATTGCAATCAGCAAGGGAGCAACTTAAGTGGTCCTTATTGAAATGA
Protein
MEGSLFEPTLYIVKGMCILDYEGNRILAKYYDKDVLPTTKEQKAFEKNLFNKTHRANAEIIMLDGLTCVYKSNVDLFFYVMGSSHENELILQSVLNALYESVSLLLRRNMERRVLMENLDAVMLAFDEICDGGVILDADPTSIVSRAALRTEDVPLGEQTVAQVLQSAREQLKWSLLK
Summary
Uniprot
D2Y4R7
Q2F5X8
I4DNA1
I4DMZ6
A0A194PUC7
A0A1E1WMW9
+ More
S4PB31 A0A212ENI6 H9JJQ9 A0A2A4JXX7 A0A2H1V7U4 A0A1E1WJ42 K7J3B5 A0A2H1VUV1 J9HTQ2 A0A023EGZ8 B0W863 A0A158NTZ6 A0A139WEJ8 A0A1L8DVP2 Q16K40 A0A3L8DN81 A0A0C9QIL8 A0A0L7RF57 U5EXW1 A0A1Y1MM58 A0A0J7KYC4 A0A067QZB9 A0A2P8XWI4 A0A0P4VTE4 A0A154P3D0 A0A182WBW8 A0A069DWL2 A0A195FL34 A0A088AC71 A0A0K8U5V1 E9IQ51 A0A0N0U2Y9 A0A182LSV6 A0A182RU48 A0A182K3P4 A0A182T9I0 A0A2A3EP29 V9IGN4 A0A195C7F4 A0A0A9WEI1 A0A182QL23 A0A1W4X0A8 A0A182VDE7 A0A182U1D6 A0A182WXB9 A0A182LKL5 Q7Q5C2 A0A182I4A8 A0A151I4P4 A0A0C9R460 E0VA85 A0A182NIW7 A0A182Y7W5 A0A182IRP8 A0A0T6AX76 A0A0K8R9L9 W8BUQ5 A0A158NTZ7 A0A0A1WQV6 A0A2R5LND1 A0A224YVH4 A0A131Z538 L7M6F0 A0A131XAT0 C4WUX3 A0A023GFJ6 A0A023FG69 A0A336MP29 A0A084WMK8 A0A0K8TSW5 T1JLE9 A0A023FTY2 E9H019 B4LG45 A0A3B0KLW4 B4N3N4 B5DRG2 E2C335 B3M4F3 B4QMW8 Q9VV89 D2NUJ2 A0A151IXJ4 F4WXA4 W5JB88 A0A1W4UUN2 A0A1A9YGX2 A0A1B0BY44 A0A0L0CCL3 A0A023GHX4 B4KXK2 A0A2M3Z7Q5 B4PKB4 B4HJR4
S4PB31 A0A212ENI6 H9JJQ9 A0A2A4JXX7 A0A2H1V7U4 A0A1E1WJ42 K7J3B5 A0A2H1VUV1 J9HTQ2 A0A023EGZ8 B0W863 A0A158NTZ6 A0A139WEJ8 A0A1L8DVP2 Q16K40 A0A3L8DN81 A0A0C9QIL8 A0A0L7RF57 U5EXW1 A0A1Y1MM58 A0A0J7KYC4 A0A067QZB9 A0A2P8XWI4 A0A0P4VTE4 A0A154P3D0 A0A182WBW8 A0A069DWL2 A0A195FL34 A0A088AC71 A0A0K8U5V1 E9IQ51 A0A0N0U2Y9 A0A182LSV6 A0A182RU48 A0A182K3P4 A0A182T9I0 A0A2A3EP29 V9IGN4 A0A195C7F4 A0A0A9WEI1 A0A182QL23 A0A1W4X0A8 A0A182VDE7 A0A182U1D6 A0A182WXB9 A0A182LKL5 Q7Q5C2 A0A182I4A8 A0A151I4P4 A0A0C9R460 E0VA85 A0A182NIW7 A0A182Y7W5 A0A182IRP8 A0A0T6AX76 A0A0K8R9L9 W8BUQ5 A0A158NTZ7 A0A0A1WQV6 A0A2R5LND1 A0A224YVH4 A0A131Z538 L7M6F0 A0A131XAT0 C4WUX3 A0A023GFJ6 A0A023FG69 A0A336MP29 A0A084WMK8 A0A0K8TSW5 T1JLE9 A0A023FTY2 E9H019 B4LG45 A0A3B0KLW4 B4N3N4 B5DRG2 E2C335 B3M4F3 B4QMW8 Q9VV89 D2NUJ2 A0A151IXJ4 F4WXA4 W5JB88 A0A1W4UUN2 A0A1A9YGX2 A0A1B0BY44 A0A0L0CCL3 A0A023GHX4 B4KXK2 A0A2M3Z7Q5 B4PKB4 B4HJR4
Pubmed
20967265
22651552
26354079
23622113
22118469
19121390
+ More
20075255 17510324 24945155 26483478 21347285 18362917 19820115 30249741 28004739 24845553 29403074 27129103 26334808 21282665 25401762 20966253 12364791 14747013 17210077 20566863 25244985 24495485 25830018 28797301 26830274 25576852 28049606 24438588 26369729 21292972 17994087 15632085 20798317 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 20920257 23761445 26108605 17550304
20075255 17510324 24945155 26483478 21347285 18362917 19820115 30249741 28004739 24845553 29403074 27129103 26334808 21282665 25401762 20966253 12364791 14747013 17210077 20566863 25244985 24495485 25830018 28797301 26830274 25576852 28049606 24438588 26369729 21292972 17994087 15632085 20798317 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 20920257 23761445 26108605 17550304
EMBL
GU322820
ADB66739.1
DQ311295
ABD36239.1
AK402796
BAM19391.1
+ More
AK402664 BAM19286.1 KQ459593 KPI96578.1 GDQN01002719 JAT88335.1 GAIX01006142 JAA86418.1 AGBW02013642 OWR43048.1 BABH01017599 NWSH01000462 PCG76242.1 ODYU01000881 SOQ36314.1 GDQN01004173 JAT86881.1 ODYU01004292 SOQ44004.1 CH477978 EJY58023.1 JXUM01000854 GAPW01004970 JAC08628.1 DS231857 EDS38688.1 ADTU01026224 KQ971354 KYB26287.1 GFDF01003601 JAV10483.1 EAT34662.1 QOIP01000006 RLU21900.1 GBYB01014593 JAG84360.1 KQ414608 KOC69463.1 GANO01000745 JAB59126.1 GEZM01027394 JAV86762.1 LBMM01002087 KMQ95288.1 KK852811 KDR15905.1 PYGN01001240 PSN36366.1 GDKW01000806 JAI55789.1 KQ434796 KZC05640.1 GBGD01003170 JAC85719.1 KQ981522 KYN40699.1 GDHF01030285 JAI22029.1 GL764659 EFZ17369.1 KQ435970 KOX67801.1 AXCM01000274 KZ288210 PBC32956.1 JR044033 AEY59832.1 KQ978143 KYM96777.1 GBHO01037440 JAG06164.1 AXCN02000476 AAAB01008960 EAA11346.4 APCN01002296 KQ976445 KYM86082.1 GBYB01011089 JAG80856.1 DS235005 EEB10291.1 LJIG01022621 KRT79588.1 GADI01006240 JAA67568.1 GAMC01005877 JAC00679.1 GBXI01012853 JAD01439.1 GGLE01006906 MBY11032.1 GFPF01006828 MAA17974.1 GEDV01002073 JAP86484.1 GACK01006301 JAA58733.1 GEFH01003797 JAP64784.1 ABLF02019447 ABLF02033882 AK341261 AK342301 BAH71693.1 BAH72381.1 GBBM01002794 JAC32624.1 GBBK01003976 GBBK01003975 JAC20507.1 UFQT01000224 UFQT01001316 SSX22031.1 SSX30007.1 ATLV01024481 KE525352 KFB51452.1 GDAI01000375 JAI17228.1 JH431739 GBBL01002039 JAC25281.1 GL732579 EFX74963.1 CH940647 EDW69353.1 OUUW01000012 SPP87569.1 CH964095 EDW79239.1 CH379070 EDY74131.1 GL452274 EFN77651.1 CH902618 EDV40447.1 CM000363 CM002912 EDX10760.1 KMZ00090.1 AE014296 AB037672 AAF49428.2 BAA90485.1 BT120135 ADB57056.1 KQ980828 KYN12417.1 GL888423 EGI61153.1 ADMH02001882 ETN60668.1 JXJN01022478 JRES01000681 KNC29199.1 GBBM01002795 JAC32623.1 CH933809 EDW18688.2 GGFM01003793 MBW24544.1 CM000159 EDW94812.1 CH480815 EDW41791.1
AK402664 BAM19286.1 KQ459593 KPI96578.1 GDQN01002719 JAT88335.1 GAIX01006142 JAA86418.1 AGBW02013642 OWR43048.1 BABH01017599 NWSH01000462 PCG76242.1 ODYU01000881 SOQ36314.1 GDQN01004173 JAT86881.1 ODYU01004292 SOQ44004.1 CH477978 EJY58023.1 JXUM01000854 GAPW01004970 JAC08628.1 DS231857 EDS38688.1 ADTU01026224 KQ971354 KYB26287.1 GFDF01003601 JAV10483.1 EAT34662.1 QOIP01000006 RLU21900.1 GBYB01014593 JAG84360.1 KQ414608 KOC69463.1 GANO01000745 JAB59126.1 GEZM01027394 JAV86762.1 LBMM01002087 KMQ95288.1 KK852811 KDR15905.1 PYGN01001240 PSN36366.1 GDKW01000806 JAI55789.1 KQ434796 KZC05640.1 GBGD01003170 JAC85719.1 KQ981522 KYN40699.1 GDHF01030285 JAI22029.1 GL764659 EFZ17369.1 KQ435970 KOX67801.1 AXCM01000274 KZ288210 PBC32956.1 JR044033 AEY59832.1 KQ978143 KYM96777.1 GBHO01037440 JAG06164.1 AXCN02000476 AAAB01008960 EAA11346.4 APCN01002296 KQ976445 KYM86082.1 GBYB01011089 JAG80856.1 DS235005 EEB10291.1 LJIG01022621 KRT79588.1 GADI01006240 JAA67568.1 GAMC01005877 JAC00679.1 GBXI01012853 JAD01439.1 GGLE01006906 MBY11032.1 GFPF01006828 MAA17974.1 GEDV01002073 JAP86484.1 GACK01006301 JAA58733.1 GEFH01003797 JAP64784.1 ABLF02019447 ABLF02033882 AK341261 AK342301 BAH71693.1 BAH72381.1 GBBM01002794 JAC32624.1 GBBK01003976 GBBK01003975 JAC20507.1 UFQT01000224 UFQT01001316 SSX22031.1 SSX30007.1 ATLV01024481 KE525352 KFB51452.1 GDAI01000375 JAI17228.1 JH431739 GBBL01002039 JAC25281.1 GL732579 EFX74963.1 CH940647 EDW69353.1 OUUW01000012 SPP87569.1 CH964095 EDW79239.1 CH379070 EDY74131.1 GL452274 EFN77651.1 CH902618 EDV40447.1 CM000363 CM002912 EDX10760.1 KMZ00090.1 AE014296 AB037672 AAF49428.2 BAA90485.1 BT120135 ADB57056.1 KQ980828 KYN12417.1 GL888423 EGI61153.1 ADMH02001882 ETN60668.1 JXJN01022478 JRES01000681 KNC29199.1 GBBM01002795 JAC32623.1 CH933809 EDW18688.2 GGFM01003793 MBW24544.1 CM000159 EDW94812.1 CH480815 EDW41791.1
Proteomes
UP000053268
UP000007151
UP000005204
UP000218220
UP000002358
UP000008820
+ More
UP000069940 UP000002320 UP000005205 UP000007266 UP000279307 UP000053825 UP000036403 UP000027135 UP000245037 UP000076502 UP000075920 UP000078541 UP000005203 UP000053105 UP000075883 UP000075900 UP000075881 UP000075901 UP000242457 UP000078542 UP000075886 UP000192223 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000078540 UP000009046 UP000075884 UP000076408 UP000075880 UP000007819 UP000030765 UP000000305 UP000008792 UP000268350 UP000007798 UP000001819 UP000008237 UP000007801 UP000000304 UP000000803 UP000078492 UP000007755 UP000000673 UP000192221 UP000092443 UP000092460 UP000037069 UP000009192 UP000002282 UP000001292
UP000069940 UP000002320 UP000005205 UP000007266 UP000279307 UP000053825 UP000036403 UP000027135 UP000245037 UP000076502 UP000075920 UP000078541 UP000005203 UP000053105 UP000075883 UP000075900 UP000075881 UP000075901 UP000242457 UP000078542 UP000075886 UP000192223 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000078540 UP000009046 UP000075884 UP000076408 UP000075880 UP000007819 UP000030765 UP000000305 UP000008792 UP000268350 UP000007798 UP000001819 UP000008237 UP000007801 UP000000304 UP000000803 UP000078492 UP000007755 UP000000673 UP000192221 UP000092443 UP000092460 UP000037069 UP000009192 UP000002282 UP000001292
Pfam
PF01217 Clat_adaptor_s
Interpro
CDD
ProteinModelPortal
D2Y4R7
Q2F5X8
I4DNA1
I4DMZ6
A0A194PUC7
A0A1E1WMW9
+ More
S4PB31 A0A212ENI6 H9JJQ9 A0A2A4JXX7 A0A2H1V7U4 A0A1E1WJ42 K7J3B5 A0A2H1VUV1 J9HTQ2 A0A023EGZ8 B0W863 A0A158NTZ6 A0A139WEJ8 A0A1L8DVP2 Q16K40 A0A3L8DN81 A0A0C9QIL8 A0A0L7RF57 U5EXW1 A0A1Y1MM58 A0A0J7KYC4 A0A067QZB9 A0A2P8XWI4 A0A0P4VTE4 A0A154P3D0 A0A182WBW8 A0A069DWL2 A0A195FL34 A0A088AC71 A0A0K8U5V1 E9IQ51 A0A0N0U2Y9 A0A182LSV6 A0A182RU48 A0A182K3P4 A0A182T9I0 A0A2A3EP29 V9IGN4 A0A195C7F4 A0A0A9WEI1 A0A182QL23 A0A1W4X0A8 A0A182VDE7 A0A182U1D6 A0A182WXB9 A0A182LKL5 Q7Q5C2 A0A182I4A8 A0A151I4P4 A0A0C9R460 E0VA85 A0A182NIW7 A0A182Y7W5 A0A182IRP8 A0A0T6AX76 A0A0K8R9L9 W8BUQ5 A0A158NTZ7 A0A0A1WQV6 A0A2R5LND1 A0A224YVH4 A0A131Z538 L7M6F0 A0A131XAT0 C4WUX3 A0A023GFJ6 A0A023FG69 A0A336MP29 A0A084WMK8 A0A0K8TSW5 T1JLE9 A0A023FTY2 E9H019 B4LG45 A0A3B0KLW4 B4N3N4 B5DRG2 E2C335 B3M4F3 B4QMW8 Q9VV89 D2NUJ2 A0A151IXJ4 F4WXA4 W5JB88 A0A1W4UUN2 A0A1A9YGX2 A0A1B0BY44 A0A0L0CCL3 A0A023GHX4 B4KXK2 A0A2M3Z7Q5 B4PKB4 B4HJR4
S4PB31 A0A212ENI6 H9JJQ9 A0A2A4JXX7 A0A2H1V7U4 A0A1E1WJ42 K7J3B5 A0A2H1VUV1 J9HTQ2 A0A023EGZ8 B0W863 A0A158NTZ6 A0A139WEJ8 A0A1L8DVP2 Q16K40 A0A3L8DN81 A0A0C9QIL8 A0A0L7RF57 U5EXW1 A0A1Y1MM58 A0A0J7KYC4 A0A067QZB9 A0A2P8XWI4 A0A0P4VTE4 A0A154P3D0 A0A182WBW8 A0A069DWL2 A0A195FL34 A0A088AC71 A0A0K8U5V1 E9IQ51 A0A0N0U2Y9 A0A182LSV6 A0A182RU48 A0A182K3P4 A0A182T9I0 A0A2A3EP29 V9IGN4 A0A195C7F4 A0A0A9WEI1 A0A182QL23 A0A1W4X0A8 A0A182VDE7 A0A182U1D6 A0A182WXB9 A0A182LKL5 Q7Q5C2 A0A182I4A8 A0A151I4P4 A0A0C9R460 E0VA85 A0A182NIW7 A0A182Y7W5 A0A182IRP8 A0A0T6AX76 A0A0K8R9L9 W8BUQ5 A0A158NTZ7 A0A0A1WQV6 A0A2R5LND1 A0A224YVH4 A0A131Z538 L7M6F0 A0A131XAT0 C4WUX3 A0A023GFJ6 A0A023FG69 A0A336MP29 A0A084WMK8 A0A0K8TSW5 T1JLE9 A0A023FTY2 E9H019 B4LG45 A0A3B0KLW4 B4N3N4 B5DRG2 E2C335 B3M4F3 B4QMW8 Q9VV89 D2NUJ2 A0A151IXJ4 F4WXA4 W5JB88 A0A1W4UUN2 A0A1A9YGX2 A0A1B0BY44 A0A0L0CCL3 A0A023GHX4 B4KXK2 A0A2M3Z7Q5 B4PKB4 B4HJR4
PDB
5NZV
E-value=3.40052e-60,
Score=582
Ontologies
GO
PANTHER
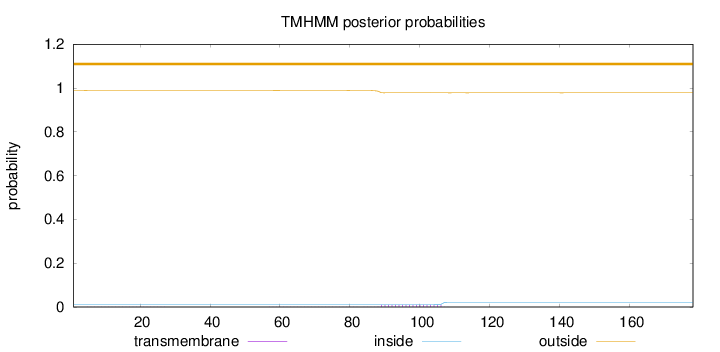
Topology
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2108
Exp number, first 60 AAs:
0.00763
Total prob of N-in:
0.01142
outside
1 - 178
Population Genetic Test Statistics
Pi
282.058471
Theta
22.754578
Tajima's D
0.723528
CLR
1.818015
CSRT
0.593870306484676
Interpretation
Uncertain
