Gene
KWMTBOMO00859
Annotation
PREDICTED:_homeotic_protein_distal-less_isoform_X2_[Bombyx_mori]
Full name
Homeotic protein distal-less
Alternative Name
Protein brista
Transcription factor
Location in the cell
Nuclear Reliability : 4.736
Sequence
CDS
ATGACATCACAGGATAACCTCGACCACCACCACCTGGGGGGCTCGCAGACCCCCCACGATATATCGAATTCAGCCAACTCGACCCCCACGAATGTGTCAAAATCTGCGTTTATAGAATTACAGCAGCACGGTTATGGGCCTTTCAAGGGGGGGTATCAGCATCCCCATCATTTCGGTAGTCCGGGGGGTCAGCAGAATCCTCACGAGGCATCAGGGTTTCCCAGTCCTAGATCATTAGGTTATCCTTTCCCTCCTATGCACCAAAACACGTATGGGTACCATTTAGGGTCTTACGCGCCGCAGTGTGCAAGTCCGCCCAAAGACGAGAAATGCGGTCTCTCGGACGATCCCGGTCTCCGCGTGAACGGCAAAGGGAAGAAGATGAGGAAGCCCCGCACAATATACTCCAGCCTTCAACTGCAACAGCTGAACAGGAGGTTCCAGAGGACGCAGTACCTCGCGCTACCAGAGCGGGCAGAGTTGGCGGCCAGCCTCGGACTAACGCAGACGCAGGTAAGTTGA
Protein
MTSQDNLDHHHLGGSQTPHDISNSANSTPTNVSKSAFIELQQHGYGPFKGGYQHPHHFGSPGGQQNPHEASGFPSPRSLGYPFPPMHQNTYGYHLGSYAPQCASPPKDEKCGLSDDPGLRVNGKGKKMRKPRTIYSSLQLQQLNRRFQRTQYLALPERAELAASLGLTQTQVS
Summary
Description
Transcription factor that plays a role in larval and adult appendage development. Specifies the identity of ventral appendages (including legs and antennae) and suppresses dorsal appendage development. Involved in patterning the distal-proximal limb axis. May control the adhesive properties of cells during limb morphogenesis. Also has a secondary role in the normal patterning of the wing margin.
Keywords
Alternative splicing
Complete proteome
Developmental protein
DNA-binding
Homeobox
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Homeotic protein distal-less
splice variant In isoform B.
splice variant In isoform B.
Uniprot
Q9TX42
A0A194R167
Q6IWM7
A0A0D3S0J9
A0A212ENJ0
Q95VX3
+ More
Q8WRT8 A0A2Z6DQ67 A0A0D3S0F4 A0A194PT33 A0A2A4J7K9 B4X6Z7 A0A2H1VT44 A0A1B6FAV6 A0A182IKW0 A0A1B6CPJ5 A0A1B6L2D8 A0A182Y262 A0A1B6LCK2 A0A182II11 Q7PTF4 A0A182NZ04 A0A182VK49 A0A182XU98 A0A1W6GWZ2 A0A0K8SKT1 A0A182L384 A0A034WJE2 A0A0K8W1B8 A0A0K8VQ10 A0A0K8WMP1 W8BZ01 A0A1J1ITQ0 B4LN64 A0A0J9RL03 A0A0Q9WFJ6 B4IH92 B3NQZ9 B4QCK9 P20009 A0A1W4V1H5 P20009-2 A2VEF7 A0A0J9RKF8 A0A0Q5VYI1 B4PBV9 A0A0R1DTV4 B4J4C5 A0A0M4EKJ2 A0A1W4V082 A0A0A1X4Q1 A0A1W4UMZ4 A0A0A1WST2 A0A1I8MVV3 A0A1I8MVV0 A0A1I8P6X7 A0A1I8P6S5 A0A0L0BPR6 B4MPA7 B4KTK2 A0A3B0JGM8 Q28Z29 B4GIC8 B3MC35 A0A336M6W2 A0A0R3NNK2 A0A1A9V9P4 A0A1W4XKR1 A0A1B0CN62 A0A1A9WBB7 A0A1I8MVU4 A0A1I8P6U2 A0A1I8P6T1 A0A1I8MVU2 A0A1A9YDK1 A0A3Q0J0V4 A0A1B0BB12 A0A2R7W6F3 N6SY71 B3IWG0 A0A1B0FKQ2 A0A0C9QHP3 D6WRE0 A0A1Y1MUN0 Q9GPL3 Q2MHJ4 A0A2S2R447 A0A154PTJ9 J9L6R3 A0A1Q2SJQ0 A0A0H5AYY2 A0A2H8TVP1 B0I561 A0A2A3EHH7 A0A087ZMZ9
Q8WRT8 A0A2Z6DQ67 A0A0D3S0F4 A0A194PT33 A0A2A4J7K9 B4X6Z7 A0A2H1VT44 A0A1B6FAV6 A0A182IKW0 A0A1B6CPJ5 A0A1B6L2D8 A0A182Y262 A0A1B6LCK2 A0A182II11 Q7PTF4 A0A182NZ04 A0A182VK49 A0A182XU98 A0A1W6GWZ2 A0A0K8SKT1 A0A182L384 A0A034WJE2 A0A0K8W1B8 A0A0K8VQ10 A0A0K8WMP1 W8BZ01 A0A1J1ITQ0 B4LN64 A0A0J9RL03 A0A0Q9WFJ6 B4IH92 B3NQZ9 B4QCK9 P20009 A0A1W4V1H5 P20009-2 A2VEF7 A0A0J9RKF8 A0A0Q5VYI1 B4PBV9 A0A0R1DTV4 B4J4C5 A0A0M4EKJ2 A0A1W4V082 A0A0A1X4Q1 A0A1W4UMZ4 A0A0A1WST2 A0A1I8MVV3 A0A1I8MVV0 A0A1I8P6X7 A0A1I8P6S5 A0A0L0BPR6 B4MPA7 B4KTK2 A0A3B0JGM8 Q28Z29 B4GIC8 B3MC35 A0A336M6W2 A0A0R3NNK2 A0A1A9V9P4 A0A1W4XKR1 A0A1B0CN62 A0A1A9WBB7 A0A1I8MVU4 A0A1I8P6U2 A0A1I8P6T1 A0A1I8MVU2 A0A1A9YDK1 A0A3Q0J0V4 A0A1B0BB12 A0A2R7W6F3 N6SY71 B3IWG0 A0A1B0FKQ2 A0A0C9QHP3 D6WRE0 A0A1Y1MUN0 Q9GPL3 Q2MHJ4 A0A2S2R447 A0A154PTJ9 J9L6R3 A0A1Q2SJQ0 A0A0H5AYY2 A0A2H8TVP1 B0I561 A0A2A3EHH7 A0A087ZMZ9
Pubmed
7953552
26354079
22118469
11797007
25244985
12364791
+ More
14747013 17210077 20966253 25348373 24495485 17994087 22936249 1358457 10731132 12537572 2564639 12537569 9303541 9778507 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 25315136 26108605 15632085 23537049 18362917 19820115 28004739 11124123 27989519 20449607
14747013 17210077 20966253 25348373 24495485 17994087 22936249 1358457 10731132 12537572 2564639 12537569 9303541 9778507 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 25315136 26108605 15632085 23537049 18362917 19820115 28004739 11124123 27989519 20449607
EMBL
KQ460890
KPJ10995.1
AY616435
AAT39558.1
KP120517
AJS19035.1
+ More
AGBW02013642 OWR43046.1 AF404110 AAK97630.1 AF404825 AAL69325.1 LC033901 BBD71106.1 KP120518 AJS19036.1 KQ459593 KPI96576.1 NWSH01002506 PCG68117.1 EF362786 ABP35643.1 ODYU01004288 SOQ44000.1 GECZ01022505 JAS47264.1 GEDC01021919 GEDC01019408 JAS15379.1 JAS17890.1 GEBQ01022176 JAT17801.1 GEBQ01018587 JAT21390.1 APCN01001490 AAAB01008807 EAA03995.4 KX449133 ARM20313.1 GBRD01012419 JAG53405.1 GAKP01004168 JAC54784.1 GDHF01007433 JAI44881.1 GDHF01011340 JAI40974.1 GDHF01000084 JAI52230.1 GAMC01008054 JAB98501.1 CVRI01000059 CRL03098.1 CH940648 EDW60068.1 CM002911 KMY96447.1 KRF79216.1 CH480838 EDW49268.1 CH954179 EDV57082.1 CM000362 EDX08625.1 KMY96446.1 S47947 AE013599 AY113370 BT030126 ABN49265.1 ACL83212.1 KMY96448.1 KQS63147.1 CM000158 EDW92613.1 KRK00604.1 CH916367 EDW01607.1 CP012524 ALC41917.1 GBXI01008185 JAD06107.1 GBXI01012727 JAD01565.1 JRES01001567 KNC21963.1 CH963849 EDW73946.1 CH933808 EDW08563.2 OUUW01000001 SPP74510.1 CM000071 EAL25786.2 CH479183 EDW36248.1 CH902619 EDV37222.2 UFQT01000543 SSX25121.1 KRT02551.1 AJWK01019842 AJWK01019843 AJWK01019844 AJWK01019845 JXJN01011270 JXJN01011271 KK854331 PTY14741.1 APGK01052158 APGK01052159 APGK01052160 APGK01052161 KB741211 ENN72734.1 AB365073 BAG49469.1 CCAG010022550 CCAG010022551 GBYB01000087 JAG69854.1 KQ971351 EFA07478.1 GEZM01023730 JAV88210.1 AF317551 AAG39634.1 AB200969 BAE78537.1 GGMS01015535 MBY84738.1 KQ435105 KZC14678.1 ABLF02037685 LC199492 BAW79361.1 LC010622 BAR93875.1 GFXV01006511 MBW18316.1 AB378321 BAG06741.1 KZ288242 PBC31253.1
AGBW02013642 OWR43046.1 AF404110 AAK97630.1 AF404825 AAL69325.1 LC033901 BBD71106.1 KP120518 AJS19036.1 KQ459593 KPI96576.1 NWSH01002506 PCG68117.1 EF362786 ABP35643.1 ODYU01004288 SOQ44000.1 GECZ01022505 JAS47264.1 GEDC01021919 GEDC01019408 JAS15379.1 JAS17890.1 GEBQ01022176 JAT17801.1 GEBQ01018587 JAT21390.1 APCN01001490 AAAB01008807 EAA03995.4 KX449133 ARM20313.1 GBRD01012419 JAG53405.1 GAKP01004168 JAC54784.1 GDHF01007433 JAI44881.1 GDHF01011340 JAI40974.1 GDHF01000084 JAI52230.1 GAMC01008054 JAB98501.1 CVRI01000059 CRL03098.1 CH940648 EDW60068.1 CM002911 KMY96447.1 KRF79216.1 CH480838 EDW49268.1 CH954179 EDV57082.1 CM000362 EDX08625.1 KMY96446.1 S47947 AE013599 AY113370 BT030126 ABN49265.1 ACL83212.1 KMY96448.1 KQS63147.1 CM000158 EDW92613.1 KRK00604.1 CH916367 EDW01607.1 CP012524 ALC41917.1 GBXI01008185 JAD06107.1 GBXI01012727 JAD01565.1 JRES01001567 KNC21963.1 CH963849 EDW73946.1 CH933808 EDW08563.2 OUUW01000001 SPP74510.1 CM000071 EAL25786.2 CH479183 EDW36248.1 CH902619 EDV37222.2 UFQT01000543 SSX25121.1 KRT02551.1 AJWK01019842 AJWK01019843 AJWK01019844 AJWK01019845 JXJN01011270 JXJN01011271 KK854331 PTY14741.1 APGK01052158 APGK01052159 APGK01052160 APGK01052161 KB741211 ENN72734.1 AB365073 BAG49469.1 CCAG010022550 CCAG010022551 GBYB01000087 JAG69854.1 KQ971351 EFA07478.1 GEZM01023730 JAV88210.1 AF317551 AAG39634.1 AB200969 BAE78537.1 GGMS01015535 MBY84738.1 KQ435105 KZC14678.1 ABLF02037685 LC199492 BAW79361.1 LC010622 BAR93875.1 GFXV01006511 MBW18316.1 AB378321 BAG06741.1 KZ288242 PBC31253.1
Proteomes
UP000053240
UP000007151
UP000053268
UP000218220
UP000075880
UP000076408
+ More
UP000075840 UP000007062 UP000075884 UP000075903 UP000076407 UP000075882 UP000183832 UP000008792 UP000001292 UP000008711 UP000000304 UP000000803 UP000192221 UP000002282 UP000001070 UP000092553 UP000095301 UP000095300 UP000037069 UP000007798 UP000009192 UP000268350 UP000001819 UP000008744 UP000007801 UP000078200 UP000192223 UP000092461 UP000091820 UP000092443 UP000079169 UP000092460 UP000019118 UP000092444 UP000007266 UP000076502 UP000007819 UP000242457 UP000005203
UP000075840 UP000007062 UP000075884 UP000075903 UP000076407 UP000075882 UP000183832 UP000008792 UP000001292 UP000008711 UP000000304 UP000000803 UP000192221 UP000002282 UP000001070 UP000092553 UP000095301 UP000095300 UP000037069 UP000007798 UP000009192 UP000268350 UP000001819 UP000008744 UP000007801 UP000078200 UP000192223 UP000092461 UP000091820 UP000092443 UP000079169 UP000092460 UP000019118 UP000092444 UP000007266 UP000076502 UP000007819 UP000242457 UP000005203
Pfam
PF00046 Homeodomain
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
Q9TX42
A0A194R167
Q6IWM7
A0A0D3S0J9
A0A212ENJ0
Q95VX3
+ More
Q8WRT8 A0A2Z6DQ67 A0A0D3S0F4 A0A194PT33 A0A2A4J7K9 B4X6Z7 A0A2H1VT44 A0A1B6FAV6 A0A182IKW0 A0A1B6CPJ5 A0A1B6L2D8 A0A182Y262 A0A1B6LCK2 A0A182II11 Q7PTF4 A0A182NZ04 A0A182VK49 A0A182XU98 A0A1W6GWZ2 A0A0K8SKT1 A0A182L384 A0A034WJE2 A0A0K8W1B8 A0A0K8VQ10 A0A0K8WMP1 W8BZ01 A0A1J1ITQ0 B4LN64 A0A0J9RL03 A0A0Q9WFJ6 B4IH92 B3NQZ9 B4QCK9 P20009 A0A1W4V1H5 P20009-2 A2VEF7 A0A0J9RKF8 A0A0Q5VYI1 B4PBV9 A0A0R1DTV4 B4J4C5 A0A0M4EKJ2 A0A1W4V082 A0A0A1X4Q1 A0A1W4UMZ4 A0A0A1WST2 A0A1I8MVV3 A0A1I8MVV0 A0A1I8P6X7 A0A1I8P6S5 A0A0L0BPR6 B4MPA7 B4KTK2 A0A3B0JGM8 Q28Z29 B4GIC8 B3MC35 A0A336M6W2 A0A0R3NNK2 A0A1A9V9P4 A0A1W4XKR1 A0A1B0CN62 A0A1A9WBB7 A0A1I8MVU4 A0A1I8P6U2 A0A1I8P6T1 A0A1I8MVU2 A0A1A9YDK1 A0A3Q0J0V4 A0A1B0BB12 A0A2R7W6F3 N6SY71 B3IWG0 A0A1B0FKQ2 A0A0C9QHP3 D6WRE0 A0A1Y1MUN0 Q9GPL3 Q2MHJ4 A0A2S2R447 A0A154PTJ9 J9L6R3 A0A1Q2SJQ0 A0A0H5AYY2 A0A2H8TVP1 B0I561 A0A2A3EHH7 A0A087ZMZ9
Q8WRT8 A0A2Z6DQ67 A0A0D3S0F4 A0A194PT33 A0A2A4J7K9 B4X6Z7 A0A2H1VT44 A0A1B6FAV6 A0A182IKW0 A0A1B6CPJ5 A0A1B6L2D8 A0A182Y262 A0A1B6LCK2 A0A182II11 Q7PTF4 A0A182NZ04 A0A182VK49 A0A182XU98 A0A1W6GWZ2 A0A0K8SKT1 A0A182L384 A0A034WJE2 A0A0K8W1B8 A0A0K8VQ10 A0A0K8WMP1 W8BZ01 A0A1J1ITQ0 B4LN64 A0A0J9RL03 A0A0Q9WFJ6 B4IH92 B3NQZ9 B4QCK9 P20009 A0A1W4V1H5 P20009-2 A2VEF7 A0A0J9RKF8 A0A0Q5VYI1 B4PBV9 A0A0R1DTV4 B4J4C5 A0A0M4EKJ2 A0A1W4V082 A0A0A1X4Q1 A0A1W4UMZ4 A0A0A1WST2 A0A1I8MVV3 A0A1I8MVV0 A0A1I8P6X7 A0A1I8P6S5 A0A0L0BPR6 B4MPA7 B4KTK2 A0A3B0JGM8 Q28Z29 B4GIC8 B3MC35 A0A336M6W2 A0A0R3NNK2 A0A1A9V9P4 A0A1W4XKR1 A0A1B0CN62 A0A1A9WBB7 A0A1I8MVU4 A0A1I8P6U2 A0A1I8P6T1 A0A1I8MVU2 A0A1A9YDK1 A0A3Q0J0V4 A0A1B0BB12 A0A2R7W6F3 N6SY71 B3IWG0 A0A1B0FKQ2 A0A0C9QHP3 D6WRE0 A0A1Y1MUN0 Q9GPL3 Q2MHJ4 A0A2S2R447 A0A154PTJ9 J9L6R3 A0A1Q2SJQ0 A0A0H5AYY2 A0A2H8TVP1 B0I561 A0A2A3EHH7 A0A087ZMZ9
PDB
4RDU
E-value=2.05651e-09,
Score=144
Ontologies
GO
GO:0043565
GO:0006355
GO:0005634
GO:0003677
GO:0006357
GO:0003700
GO:0000978
GO:0021553
GO:0016319
GO:0048728
GO:0007449
GO:0042048
GO:0045893
GO:0008587
GO:0007469
GO:0007480
GO:0035114
GO:0007479
GO:0007485
GO:0035215
GO:0010092
GO:0010629
GO:0048264
GO:0007487
GO:0016021
GO:0006741
GO:0005737
GO:0006396
GO:0000166
GO:0004887
GO:0003707
Topology
Subcellular location
Nucleus
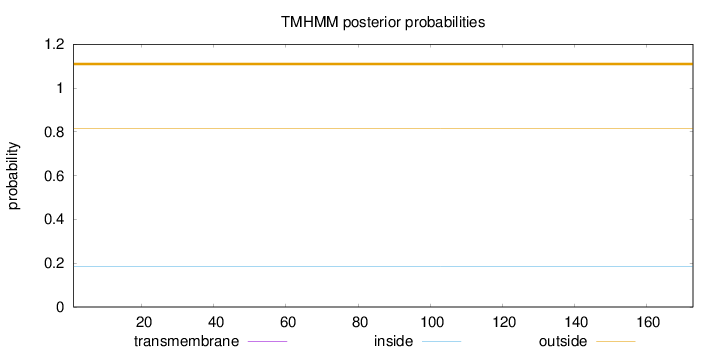
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00055
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18552
outside
1 - 173
Population Genetic Test Statistics
Pi
232.669545
Theta
173.953629
Tajima's D
1.389706
CLR
0.138862
CSRT
0.760861956902155
Interpretation
Uncertain
