Gene
KWMTBOMO00852
Pre Gene Modal
BGIBMGA009675
Annotation
PREDICTED:_4-coumarate--CoA_ligase_1-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.017
Sequence
CDS
ATGCTGAAGAATCCTAACTACGTGTACGGGTCCAGGGACATCACCGTTCCGCTGCACTTGAACTTCGGCCAGTTCATACTGGACAGAATGTGGATGCACAAAGACAGGATTGCACAGACAAATGGAGCCACAGGCGAAACTATGACCTACGGGGAAATTGTGCAGCAATCTATGAGATTCGCTATCTCGCTCTCTCGACTGGGCGTACGTAAAGGAGAGAGAATAGCGATACTATCTGAGAACAGACAAGAGTTTTGGACTGCTGTGATTGGTCCGGCATGCGTCGGCGCCATTGTCACTACTATAAGCACTAATTATGTTAAAGGCGAACTAAAACACGCAATGAACATATCAAAACCGAAATACGTGATAATATCGCCGAAAATGCACGAGAGGCACGTGCGAACGTTAAAATCGTTTGGATTCATTGAGAAAACGATAATTTTCGGCGACGCTGGGTCGAAAAGCGGTTTATATTCGTTCGACGAGTTAGTATTGGCCGGTGGCGGGTCTGAGATTGAAATCGAGAAGTTCACCGCGGTCAACGTGAATGGGCAGAACGATGTTTTGTTCATGGTCTATTCCTCTGGGACCACCGGCTTGCCAAAGGGAGTGCTGTTAACGCATTACAACGTTATAGCAGCGTGTTGTTTGCCTGTGACTTTCGACCCGAAGCAGCCGACTCTAAGCGTCGGTCCGTGGTACCACGTAATGGGTCTGGTCGGTAGCATCACTGGGATGGCTAGCGGTAGAATCGGGGTTTATCTGGACAAATTCGACGTTGAACTATACTTGAAGACAATCGAGAGGTATAAGGTGGTCCAGCTGACCGTGGTACCCCCGATCTTGGTCGCAGTCTGCAAGTCTACCAGCACTTTGGACCTGGGCTCTGTTAGACTGATATATTCGGGGGCCGCTCCCCTGCACAAGGACACGGCCAAGGCTGCCTTCGAAAAGTTCCCGAACCTTAGAGGAGTCTTCCAAGGTTACGGGTCGTCAGAGACGACCCTGGGCATCACCAGATACACGTACACCCAATTAGACATCGCGAAACCGGGCAGCGTGGGTCTGGCTGCACCCGGATGCGTGCTGAAGATAGTCGACATAAAAACCCGCCAGCCTCTTGGCCCTAATCAAACAGGAGAGATTTGCGCAAAGGGTGCGCTCCTTATGAAGGGCTACATCGGGAGGGCCAGGTCCGAAGACTTTGATGACGAGGACTTCTACAGGACTGGCGACGTAGGCTACTACGACGAGGACGGACACTTCTACATCTGTGATAGGATTAAGGAACTCATCAAATATAAAGGATATCAGGTGCCTCCAGCCGAACTGGAAGCGGTCCTGCTGCAGCACGAAGCAGTGAAAGACGCCGGTGTCGTCGGCATAGAGGACCGCGCGGCAGGAGAGGTGCCATTAGCCTTCGTTGTACGCCAACCAGGGAAGGAAGTCACTGAACAGGACCTCAAAGATTTCGTGGCACGGACGCTATCAAATCCGAAACACCTTCGGGGCGGAGTACGCTTCGTGACCGAAATACCGAAGAACCCCAGCGGGAAAATACTGAGGCAGGAGCTCAGGAAATTAGCTAAATCCCGAAAGAGCAAACTTTAA
Protein
MLKNPNYVYGSRDITVPLHLNFGQFILDRMWMHKDRIAQTNGATGETMTYGEIVQQSMRFAISLSRLGVRKGERIAILSENRQEFWTAVIGPACVGAIVTTISTNYVKGELKHAMNISKPKYVIISPKMHERHVRTLKSFGFIEKTIIFGDAGSKSGLYSFDELVLAGGGSEIEIEKFTAVNVNGQNDVLFMVYSSGTTGLPKGVLLTHYNVIAACCLPVTFDPKQPTLSVGPWYHVMGLVGSITGMASGRIGVYLDKFDVELYLKTIERYKVVQLTVVPPILVAVCKSTSTLDLGSVRLIYSGAAPLHKDTAKAAFEKFPNLRGVFQGYGSSETTLGITRYTYTQLDIAKPGSVGLAAPGCVLKIVDIKTRQPLGPNQTGEICAKGALLMKGYIGRARSEDFDDEDFYRTGDVGYYDEDGHFYICDRIKELIKYKGYQVPPAELEAVLLQHEAVKDAGVVGIEDRAAGEVPLAFVVRQPGKEVTEQDLKDFVARTLSNPKHLRGGVRFVTEIPKNPSGKILRQELRKLAKSRKSKL
Summary
Uniprot
H9JJH4
A0A2A4JYH7
A0A194R547
A0A2H1VNT1
A0A194QZV5
A0A194PZH5
+ More
A0A194QZM0 A0A194PT23 I4DPZ6 A0A212FII9 A0A212FIH9 A0A194PT28 A0A194R162 A0A194R0D8 A0A194PUB9 H9JJH5 A0A194QRL9 G8GE17 A0A2H1VTP5 H9J2F6 A0A194PHR8 A0A2A4J232 A0A194PPL0 A0A2A4JMM7 A0A2H1WNI5 A0A212EHJ8 A0A2A4JNM0 A0A2W1BTP3 A0A0C6DRW5 A0A2A4J3S6 A0A194RLN8 A0A194PGU6 A0A212ETT6 A0A2H1V0T1 A0A1L8DYF7 E2AA43 A0A1L8DYB2 A0A084W803 A0A1L8DZF9 A0A182F8R8 A0A2M4AKM3 A0A182SCF1 A0A182TMS7 A0A1L8DYZ5 A0A182IVA5 W5JL40 A0A0C6E621 A0A182FB05 A0A2M4AK32 A0A182PST3 D2KV87 A0A182YEP9 A0A182SUG3 A0A182T271 A0A182R867 A0A182WW22 A0A0L7L784 A0A182YH13 A0A1I8JVD6 A0A195CJ24 A0A182V899 U5EXU9 A0A182RE74 A7US60 A0A182HJY0 A0A2M4BJ94 A0A154P2Q1 A0A0J7KT57 A0A182MJF5 A0A182PUM4 A0A182KX68 W5JAQ2 A0A1I8JVX6 A0A158NKN5 A0A2M3Z9G4 A0A1Q3FIY4 A0A182YM72 A0A088A0E6 A0A182VS70 A0A1B0DFB0 A0A1I8JV25 A0A1I8JTC1 A0A240PK82 A0A0A1X815 A0A0L7LP26 A0A084VAF2 A0A182G4E6 A0A2A3EQ70 A0A1L8EBZ5 A0A182R329 A0A1L8ECA8 A0A182HDU2 A0A1A9XI15 F5HJ76 A0A1I8PLP9 B0W9Q1 Q17Q46 A0A1A9VQZ3 A0A182YM73
A0A194QZM0 A0A194PT23 I4DPZ6 A0A212FII9 A0A212FIH9 A0A194PT28 A0A194R162 A0A194R0D8 A0A194PUB9 H9JJH5 A0A194QRL9 G8GE17 A0A2H1VTP5 H9J2F6 A0A194PHR8 A0A2A4J232 A0A194PPL0 A0A2A4JMM7 A0A2H1WNI5 A0A212EHJ8 A0A2A4JNM0 A0A2W1BTP3 A0A0C6DRW5 A0A2A4J3S6 A0A194RLN8 A0A194PGU6 A0A212ETT6 A0A2H1V0T1 A0A1L8DYF7 E2AA43 A0A1L8DYB2 A0A084W803 A0A1L8DZF9 A0A182F8R8 A0A2M4AKM3 A0A182SCF1 A0A182TMS7 A0A1L8DYZ5 A0A182IVA5 W5JL40 A0A0C6E621 A0A182FB05 A0A2M4AK32 A0A182PST3 D2KV87 A0A182YEP9 A0A182SUG3 A0A182T271 A0A182R867 A0A182WW22 A0A0L7L784 A0A182YH13 A0A1I8JVD6 A0A195CJ24 A0A182V899 U5EXU9 A0A182RE74 A7US60 A0A182HJY0 A0A2M4BJ94 A0A154P2Q1 A0A0J7KT57 A0A182MJF5 A0A182PUM4 A0A182KX68 W5JAQ2 A0A1I8JVX6 A0A158NKN5 A0A2M3Z9G4 A0A1Q3FIY4 A0A182YM72 A0A088A0E6 A0A182VS70 A0A1B0DFB0 A0A1I8JV25 A0A1I8JTC1 A0A240PK82 A0A0A1X815 A0A0L7LP26 A0A084VAF2 A0A182G4E6 A0A2A3EQ70 A0A1L8EBZ5 A0A182R329 A0A1L8ECA8 A0A182HDU2 A0A1A9XI15 F5HJ76 A0A1I8PLP9 B0W9Q1 Q17Q46 A0A1A9VQZ3 A0A182YM73
Pubmed
EMBL
BABH01017566
BABH01017567
NWSH01000380
PCG76826.1
KQ460890
KPJ10986.1
+ More
ODYU01003402 SOQ42112.1 KPJ10987.1 KQ459593 KPI96565.1 KPJ10988.1 KPI96566.1 AK403861 BAM19986.1 AGBW02008380 OWR53544.1 OWR53543.1 KPI96571.1 KPJ10990.1 KPJ10989.1 KPI96568.1 BABH01017564 KQ461181 KPJ07625.1 JN127350 AET05796.1 ODYU01004352 SOQ44136.1 BABH01007891 KQ459604 KPI92264.1 NWSH01003686 PCG65919.1 KQ459597 KPI94913.1 NWSH01001030 PCG73036.1 ODYU01009511 SOQ54004.1 AGBW02014874 OWR40957.1 PCG73033.1 KZ149903 PZC78438.1 AB251887 BAQ25864.1 NWSH01003607 PCG66062.1 KQ460045 KPJ18230.1 KPI92263.1 AGBW02012531 OWR44889.1 ODYU01000123 SOQ34366.1 GFDF01002635 JAV11449.1 GL438029 EFN69681.1 GFDF01002634 JAV11450.1 ATLV01021336 KE525316 KFB46347.1 GFDF01002419 JAV11665.1 GGFK01007847 MBW41168.1 GFDF01002420 JAV11664.1 ADMH02001214 ETN63630.1 AB251886 BAQ25863.1 GGFK01007825 MBW41146.1 AB479114 BAI66602.1 JTDY01002474 KOB71343.1 KQ977649 KYN00718.1 GANO01000766 JAB59105.1 AAAB01008820 EDO64449.2 APCN01002221 GGFJ01003984 MBW53125.1 KQ434809 KZC06219.1 LBMM01003413 KMQ93562.1 AXCM01006981 ADMH02001657 ETN61532.1 ADTU01018928 GGFM01004391 MBW25142.1 GFDL01007475 JAV27570.1 AJVK01015100 AJVK01015101 AJVK01015102 AJVK01015103 APCN01002222 GBXI01007236 JAD07056.1 JTDY01000482 KOB76976.1 ATLV01003206 KE524124 KFB34946.1 JXUM01143620 KQ569633 KXJ68569.1 KZ288194 PBC33943.1 GFDG01002539 JAV16260.1 GFDG01002537 JAV16262.1 JXUM01130360 KQ567568 KXJ69363.1 EGK96337.1 DS231865 EDS40370.1 CH477187 EAT48870.1
ODYU01003402 SOQ42112.1 KPJ10987.1 KQ459593 KPI96565.1 KPJ10988.1 KPI96566.1 AK403861 BAM19986.1 AGBW02008380 OWR53544.1 OWR53543.1 KPI96571.1 KPJ10990.1 KPJ10989.1 KPI96568.1 BABH01017564 KQ461181 KPJ07625.1 JN127350 AET05796.1 ODYU01004352 SOQ44136.1 BABH01007891 KQ459604 KPI92264.1 NWSH01003686 PCG65919.1 KQ459597 KPI94913.1 NWSH01001030 PCG73036.1 ODYU01009511 SOQ54004.1 AGBW02014874 OWR40957.1 PCG73033.1 KZ149903 PZC78438.1 AB251887 BAQ25864.1 NWSH01003607 PCG66062.1 KQ460045 KPJ18230.1 KPI92263.1 AGBW02012531 OWR44889.1 ODYU01000123 SOQ34366.1 GFDF01002635 JAV11449.1 GL438029 EFN69681.1 GFDF01002634 JAV11450.1 ATLV01021336 KE525316 KFB46347.1 GFDF01002419 JAV11665.1 GGFK01007847 MBW41168.1 GFDF01002420 JAV11664.1 ADMH02001214 ETN63630.1 AB251886 BAQ25863.1 GGFK01007825 MBW41146.1 AB479114 BAI66602.1 JTDY01002474 KOB71343.1 KQ977649 KYN00718.1 GANO01000766 JAB59105.1 AAAB01008820 EDO64449.2 APCN01002221 GGFJ01003984 MBW53125.1 KQ434809 KZC06219.1 LBMM01003413 KMQ93562.1 AXCM01006981 ADMH02001657 ETN61532.1 ADTU01018928 GGFM01004391 MBW25142.1 GFDL01007475 JAV27570.1 AJVK01015100 AJVK01015101 AJVK01015102 AJVK01015103 APCN01002222 GBXI01007236 JAD07056.1 JTDY01000482 KOB76976.1 ATLV01003206 KE524124 KFB34946.1 JXUM01143620 KQ569633 KXJ68569.1 KZ288194 PBC33943.1 GFDG01002539 JAV16260.1 GFDG01002537 JAV16262.1 JXUM01130360 KQ567568 KXJ69363.1 EGK96337.1 DS231865 EDS40370.1 CH477187 EAT48870.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000000311
+ More
UP000030765 UP000069272 UP000075901 UP000075902 UP000075880 UP000000673 UP000075885 UP000076408 UP000075900 UP000076407 UP000037510 UP000075903 UP000078542 UP000007062 UP000075840 UP000076502 UP000036403 UP000075883 UP000075882 UP000005205 UP000005203 UP000075920 UP000092462 UP000069940 UP000249989 UP000242457 UP000092443 UP000095300 UP000002320 UP000008820 UP000078200
UP000030765 UP000069272 UP000075901 UP000075902 UP000075880 UP000000673 UP000075885 UP000076408 UP000075900 UP000076407 UP000037510 UP000075903 UP000078542 UP000007062 UP000075840 UP000076502 UP000036403 UP000075883 UP000075882 UP000005205 UP000005203 UP000075920 UP000092462 UP000069940 UP000249989 UP000242457 UP000092443 UP000095300 UP000002320 UP000008820 UP000078200
Interpro
Gene 3D
ProteinModelPortal
H9JJH4
A0A2A4JYH7
A0A194R547
A0A2H1VNT1
A0A194QZV5
A0A194PZH5
+ More
A0A194QZM0 A0A194PT23 I4DPZ6 A0A212FII9 A0A212FIH9 A0A194PT28 A0A194R162 A0A194R0D8 A0A194PUB9 H9JJH5 A0A194QRL9 G8GE17 A0A2H1VTP5 H9J2F6 A0A194PHR8 A0A2A4J232 A0A194PPL0 A0A2A4JMM7 A0A2H1WNI5 A0A212EHJ8 A0A2A4JNM0 A0A2W1BTP3 A0A0C6DRW5 A0A2A4J3S6 A0A194RLN8 A0A194PGU6 A0A212ETT6 A0A2H1V0T1 A0A1L8DYF7 E2AA43 A0A1L8DYB2 A0A084W803 A0A1L8DZF9 A0A182F8R8 A0A2M4AKM3 A0A182SCF1 A0A182TMS7 A0A1L8DYZ5 A0A182IVA5 W5JL40 A0A0C6E621 A0A182FB05 A0A2M4AK32 A0A182PST3 D2KV87 A0A182YEP9 A0A182SUG3 A0A182T271 A0A182R867 A0A182WW22 A0A0L7L784 A0A182YH13 A0A1I8JVD6 A0A195CJ24 A0A182V899 U5EXU9 A0A182RE74 A7US60 A0A182HJY0 A0A2M4BJ94 A0A154P2Q1 A0A0J7KT57 A0A182MJF5 A0A182PUM4 A0A182KX68 W5JAQ2 A0A1I8JVX6 A0A158NKN5 A0A2M3Z9G4 A0A1Q3FIY4 A0A182YM72 A0A088A0E6 A0A182VS70 A0A1B0DFB0 A0A1I8JV25 A0A1I8JTC1 A0A240PK82 A0A0A1X815 A0A0L7LP26 A0A084VAF2 A0A182G4E6 A0A2A3EQ70 A0A1L8EBZ5 A0A182R329 A0A1L8ECA8 A0A182HDU2 A0A1A9XI15 F5HJ76 A0A1I8PLP9 B0W9Q1 Q17Q46 A0A1A9VQZ3 A0A182YM73
A0A194QZM0 A0A194PT23 I4DPZ6 A0A212FII9 A0A212FIH9 A0A194PT28 A0A194R162 A0A194R0D8 A0A194PUB9 H9JJH5 A0A194QRL9 G8GE17 A0A2H1VTP5 H9J2F6 A0A194PHR8 A0A2A4J232 A0A194PPL0 A0A2A4JMM7 A0A2H1WNI5 A0A212EHJ8 A0A2A4JNM0 A0A2W1BTP3 A0A0C6DRW5 A0A2A4J3S6 A0A194RLN8 A0A194PGU6 A0A212ETT6 A0A2H1V0T1 A0A1L8DYF7 E2AA43 A0A1L8DYB2 A0A084W803 A0A1L8DZF9 A0A182F8R8 A0A2M4AKM3 A0A182SCF1 A0A182TMS7 A0A1L8DYZ5 A0A182IVA5 W5JL40 A0A0C6E621 A0A182FB05 A0A2M4AK32 A0A182PST3 D2KV87 A0A182YEP9 A0A182SUG3 A0A182T271 A0A182R867 A0A182WW22 A0A0L7L784 A0A182YH13 A0A1I8JVD6 A0A195CJ24 A0A182V899 U5EXU9 A0A182RE74 A7US60 A0A182HJY0 A0A2M4BJ94 A0A154P2Q1 A0A0J7KT57 A0A182MJF5 A0A182PUM4 A0A182KX68 W5JAQ2 A0A1I8JVX6 A0A158NKN5 A0A2M3Z9G4 A0A1Q3FIY4 A0A182YM72 A0A088A0E6 A0A182VS70 A0A1B0DFB0 A0A1I8JV25 A0A1I8JTC1 A0A240PK82 A0A0A1X815 A0A0L7LP26 A0A084VAF2 A0A182G4E6 A0A2A3EQ70 A0A1L8EBZ5 A0A182R329 A0A1L8ECA8 A0A182HDU2 A0A1A9XI15 F5HJ76 A0A1I8PLP9 B0W9Q1 Q17Q46 A0A1A9VQZ3 A0A182YM73
PDB
2D1S
E-value=7.66234e-92,
Score=861
Ontologies
GO
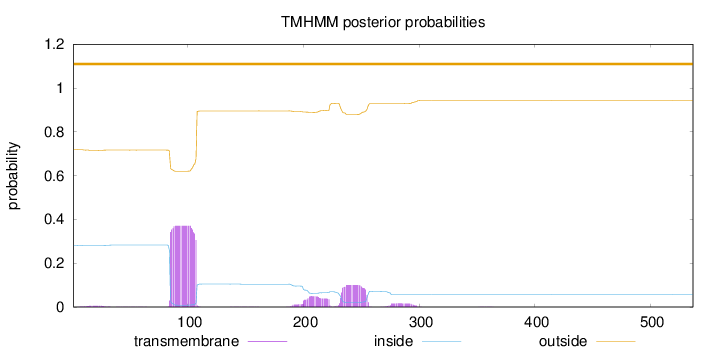
Topology
Length:
537
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.09553
Exp number, first 60 AAs:
0.09768
Total prob of N-in:
0.28144
outside
1 - 537
Population Genetic Test Statistics
Pi
301.129104
Theta
169.945384
Tajima's D
3.109858
CLR
0.016898
CSRT
0.983650817459127
Interpretation
Uncertain
