Gene
KWMTBOMO00851
Pre Gene Modal
BGIBMGA009676
Annotation
PREDICTED:_4-coumarate--CoA_ligase_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.081
Sequence
CDS
ATGCTGAAGATACCTAATTACGTGTACGGGCCCAAGGATACCACAATCCCTCTAGACGTTAACTTCGGCCAGTTCATACTGGGAAGAATGTGGATGTACAAAGACAGGATTGCACAGACGAACGGAGCCACAGGCGAAAGTTTTACTTACAAAGAAATCGTCCAGCAATCTATGAAATTCGCCGTCTCGCTTACTCGGCTTGGCGTGCGTAAGGGAGAGACGATAGCTCTATTTTCTGAGAACAGAAAAGAGTATTGGCCTGCCGTTATTGGGACGGTGTGTGTTGGCGCCATTTGTACTACTATAAGTAGTATGTATGCTGAAGAAGAAATAAAACACGTATTGCACATTTCGAAGCCGAAATATTTAATAGTTTCGCCTAAAATGTACGAAGCGCACGAGGCGACGTTGAAGTTAATCGATTCTATAGAGACAACCATTTTATTTGGCGACGATGCGAAAAGACCCGCTCTATATTATTTCAACGATTTAGTTGCGAGTGTCGGCTCAGATATCGATGTTGAGAATTTCATCGCAGCCGACGTTGATGTGCACGACGTTTTGTTCATACTGTATTCTTCGGGAACCACCGGCATGCCGAAAGGAGTGACATTAACTCATTTAAACGCCATAGCTGTTTGTTGTATGTGA
Protein
MLKIPNYVYGPKDTTIPLDVNFGQFILGRMWMYKDRIAQTNGATGESFTYKEIVQQSMKFAVSLTRLGVRKGETIALFSENRKEYWPAVIGTVCVGAICTTISSMYAEEEIKHVLHISKPKYLIVSPKMYEAHEATLKLIDSIETTILFGDDAKRPALYYFNDLVASVGSDIDVENFIAADVDVHDVLFILYSSGTTGMPKGVTLTHLNAIAVCCM
Summary
Uniprot
H9JJH4
A0A2A4JYH7
A0A2H1WXP8
A0A2H1VNT1
A0A194QZV5
A0A194R547
+ More
A0A194PZH5 A0A194R0D8 A0A194PTD5 A0A212FII9 A0A194PT23 A0A194QZM0 I4DPZ6 A0A212FIH9 A0A194R162 A0A194PUB9 A0A0L7L8I0 A0A194PUC3 A0A194PT28 A0A2A4J3S6 A0A2A4JNM0 A0A212ETT6 A0A2H1V0T1 G8GE17 A0A0L7L784 A0A2H1VTP5 H9JXM0 A0A2A4J232 A0A2H1WNI5 A0A0L7LP26 A0A0L7L6L8 H9J2F6 A0A194QRL9 A0A194RLN8 A0A194PGU6 A0A2H1VKC8 A0A0L7L765 A0A2H1V0C4 A0A2H1WJN9 A0A194PHR8 A0A2A4JMM7 A0A2W1BTP3 A0A2P8YFR7 A0A212EHJ8 A0A194PPL0 H9J5Y1 A0A0L7LH10 A0A0N0BDE6 Q17Q46 A0A0L7R1M1 A0A1S4EV05 A0A088A0E6 A0A2A3EQ70 A0A154P2Q1 K7J1G6 A0A195FCB7 A0A2C9GG62 A0A3M1BMG2 A0A158NKN5 A0A0M8K9R9 A0A0P6YM72 J9HHI9 A0A232FI37 A0A151WRK1 B3RJA0 A0A1B6G7E7 A0A1S3HPH1 A0A195DG47 A0A182PUM4 A0A369RUA1 A0A195CJ24 A0A1B6GNG2 A0A2B4R799 A0A1S3ID49 A0A182G4E6 Q7PSL0 B3RXG7 A0A182YM72 A0A1L8DZF9 A0A182R867 A0A369RY28 A0A182SCF1 A0A1Q3FIY4 A0A2J7QLS2 A0A2P8Y7W7 A0A182WW22 A0A2C9KWG2 A0A1S3J1W1 A0A182QR12 A0A1L8DYZ5 A0A1S3J1C6 A0A182V899 A0A2J7QLS4 A0A182FB05 A0A182TRT7 A0A182TMS7 F5HJ76
A0A194PZH5 A0A194R0D8 A0A194PTD5 A0A212FII9 A0A194PT23 A0A194QZM0 I4DPZ6 A0A212FIH9 A0A194R162 A0A194PUB9 A0A0L7L8I0 A0A194PUC3 A0A194PT28 A0A2A4J3S6 A0A2A4JNM0 A0A212ETT6 A0A2H1V0T1 G8GE17 A0A0L7L784 A0A2H1VTP5 H9JXM0 A0A2A4J232 A0A2H1WNI5 A0A0L7LP26 A0A0L7L6L8 H9J2F6 A0A194QRL9 A0A194RLN8 A0A194PGU6 A0A2H1VKC8 A0A0L7L765 A0A2H1V0C4 A0A2H1WJN9 A0A194PHR8 A0A2A4JMM7 A0A2W1BTP3 A0A2P8YFR7 A0A212EHJ8 A0A194PPL0 H9J5Y1 A0A0L7LH10 A0A0N0BDE6 Q17Q46 A0A0L7R1M1 A0A1S4EV05 A0A088A0E6 A0A2A3EQ70 A0A154P2Q1 K7J1G6 A0A195FCB7 A0A2C9GG62 A0A3M1BMG2 A0A158NKN5 A0A0M8K9R9 A0A0P6YM72 J9HHI9 A0A232FI37 A0A151WRK1 B3RJA0 A0A1B6G7E7 A0A1S3HPH1 A0A195DG47 A0A182PUM4 A0A369RUA1 A0A195CJ24 A0A1B6GNG2 A0A2B4R799 A0A1S3ID49 A0A182G4E6 Q7PSL0 B3RXG7 A0A182YM72 A0A1L8DZF9 A0A182R867 A0A369RY28 A0A182SCF1 A0A1Q3FIY4 A0A2J7QLS2 A0A2P8Y7W7 A0A182WW22 A0A2C9KWG2 A0A1S3J1W1 A0A182QR12 A0A1L8DYZ5 A0A1S3J1C6 A0A182V899 A0A2J7QLS4 A0A182FB05 A0A182TRT7 A0A182TMS7 F5HJ76
Pubmed
EMBL
BABH01017566
BABH01017567
NWSH01000380
PCG76826.1
ODYU01011856
SOQ57839.1
+ More
ODYU01003402 SOQ42112.1 KQ460890 KPJ10987.1 KPJ10986.1 KQ459593 KPI96565.1 KPJ10989.1 KPI96567.1 AGBW02008380 OWR53544.1 KPI96566.1 KPJ10988.1 AK403861 BAM19986.1 OWR53543.1 KPJ10990.1 KPI96568.1 JTDY01002337 KOB71639.1 KPI96573.1 KPI96571.1 NWSH01003607 PCG66062.1 NWSH01001030 PCG73033.1 AGBW02012531 OWR44889.1 ODYU01000123 SOQ34366.1 JN127350 AET05796.1 JTDY01002474 KOB71343.1 ODYU01004352 SOQ44136.1 BABH01044598 NWSH01003686 PCG65919.1 ODYU01009511 SOQ54004.1 JTDY01000482 KOB76976.1 JTDY01002678 KOB70956.1 BABH01007891 KQ461181 KPJ07625.1 KQ460045 KPJ18230.1 KQ459604 KPI92263.1 ODYU01002875 SOQ40902.1 KOB71342.1 ODYU01000100 SOQ34295.1 ODYU01009054 SOQ53186.1 KPI92264.1 PCG73036.1 KZ149903 PZC78438.1 PYGN01000629 PSN43108.1 AGBW02014874 OWR40957.1 KQ459597 KPI94913.1 BABH01029976 JTDY01001219 KOB74496.1 KQ435863 KOX70354.1 CH477187 EAT48870.1 KQ414667 KOC64742.1 KZ288194 PBC33943.1 KQ434809 KZC06219.1 KQ981693 KYN37847.1 RFKN01000393 RME09731.1 ADTU01018928 BBZA01000298 GAP64655.1 LGKN01000009 KPL86356.1 EJY57324.1 NNAY01000193 OXU30119.1 KQ982805 KYQ50532.1 DS985241 EDV29294.1 GECZ01026909 GECZ01011404 JAS42860.1 JAS58365.1 KQ980886 KYN11865.1 NOWV01000186 RDD38267.1 KQ977649 KYN00718.1 GECZ01005818 JAS63951.1 LSMT01000817 PFX14224.1 JXUM01143620 KQ569633 KXJ68569.1 AAAB01008820 EAA05417.6 DS985245 EDV24421.1 GFDF01002419 JAV11665.1 NOWV01000134 RDD39450.1 GFDL01007475 JAV27570.1 NEVH01013241 PNF29541.1 PYGN01000822 PSN40352.1 AXCN02000092 GFDF01002420 JAV11664.1 PNF29544.1 EGK96337.1
ODYU01003402 SOQ42112.1 KQ460890 KPJ10987.1 KPJ10986.1 KQ459593 KPI96565.1 KPJ10989.1 KPI96567.1 AGBW02008380 OWR53544.1 KPI96566.1 KPJ10988.1 AK403861 BAM19986.1 OWR53543.1 KPJ10990.1 KPI96568.1 JTDY01002337 KOB71639.1 KPI96573.1 KPI96571.1 NWSH01003607 PCG66062.1 NWSH01001030 PCG73033.1 AGBW02012531 OWR44889.1 ODYU01000123 SOQ34366.1 JN127350 AET05796.1 JTDY01002474 KOB71343.1 ODYU01004352 SOQ44136.1 BABH01044598 NWSH01003686 PCG65919.1 ODYU01009511 SOQ54004.1 JTDY01000482 KOB76976.1 JTDY01002678 KOB70956.1 BABH01007891 KQ461181 KPJ07625.1 KQ460045 KPJ18230.1 KQ459604 KPI92263.1 ODYU01002875 SOQ40902.1 KOB71342.1 ODYU01000100 SOQ34295.1 ODYU01009054 SOQ53186.1 KPI92264.1 PCG73036.1 KZ149903 PZC78438.1 PYGN01000629 PSN43108.1 AGBW02014874 OWR40957.1 KQ459597 KPI94913.1 BABH01029976 JTDY01001219 KOB74496.1 KQ435863 KOX70354.1 CH477187 EAT48870.1 KQ414667 KOC64742.1 KZ288194 PBC33943.1 KQ434809 KZC06219.1 KQ981693 KYN37847.1 RFKN01000393 RME09731.1 ADTU01018928 BBZA01000298 GAP64655.1 LGKN01000009 KPL86356.1 EJY57324.1 NNAY01000193 OXU30119.1 KQ982805 KYQ50532.1 DS985241 EDV29294.1 GECZ01026909 GECZ01011404 JAS42860.1 JAS58365.1 KQ980886 KYN11865.1 NOWV01000186 RDD38267.1 KQ977649 KYN00718.1 GECZ01005818 JAS63951.1 LSMT01000817 PFX14224.1 JXUM01143620 KQ569633 KXJ68569.1 AAAB01008820 EAA05417.6 DS985245 EDV24421.1 GFDF01002419 JAV11665.1 NOWV01000134 RDD39450.1 GFDL01007475 JAV27570.1 NEVH01013241 PNF29541.1 PYGN01000822 PSN40352.1 AXCN02000092 GFDF01002420 JAV11664.1 PNF29544.1 EGK96337.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000245037 UP000053105 UP000008820 UP000053825 UP000005203 UP000242457 UP000076502 UP000002358 UP000078541 UP000005205 UP000037784 UP000050502 UP000215335 UP000075809 UP000009022 UP000085678 UP000078492 UP000075885 UP000253843 UP000078542 UP000225706 UP000069940 UP000249989 UP000007062 UP000076408 UP000075900 UP000075901 UP000235965 UP000076407 UP000076420 UP000075886 UP000075903 UP000069272 UP000075902
UP000245037 UP000053105 UP000008820 UP000053825 UP000005203 UP000242457 UP000076502 UP000002358 UP000078541 UP000005205 UP000037784 UP000050502 UP000215335 UP000075809 UP000009022 UP000085678 UP000078492 UP000075885 UP000253843 UP000078542 UP000225706 UP000069940 UP000249989 UP000007062 UP000076408 UP000075900 UP000075901 UP000235965 UP000076407 UP000076420 UP000075886 UP000075903 UP000069272 UP000075902
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
H9JJH4
A0A2A4JYH7
A0A2H1WXP8
A0A2H1VNT1
A0A194QZV5
A0A194R547
+ More
A0A194PZH5 A0A194R0D8 A0A194PTD5 A0A212FII9 A0A194PT23 A0A194QZM0 I4DPZ6 A0A212FIH9 A0A194R162 A0A194PUB9 A0A0L7L8I0 A0A194PUC3 A0A194PT28 A0A2A4J3S6 A0A2A4JNM0 A0A212ETT6 A0A2H1V0T1 G8GE17 A0A0L7L784 A0A2H1VTP5 H9JXM0 A0A2A4J232 A0A2H1WNI5 A0A0L7LP26 A0A0L7L6L8 H9J2F6 A0A194QRL9 A0A194RLN8 A0A194PGU6 A0A2H1VKC8 A0A0L7L765 A0A2H1V0C4 A0A2H1WJN9 A0A194PHR8 A0A2A4JMM7 A0A2W1BTP3 A0A2P8YFR7 A0A212EHJ8 A0A194PPL0 H9J5Y1 A0A0L7LH10 A0A0N0BDE6 Q17Q46 A0A0L7R1M1 A0A1S4EV05 A0A088A0E6 A0A2A3EQ70 A0A154P2Q1 K7J1G6 A0A195FCB7 A0A2C9GG62 A0A3M1BMG2 A0A158NKN5 A0A0M8K9R9 A0A0P6YM72 J9HHI9 A0A232FI37 A0A151WRK1 B3RJA0 A0A1B6G7E7 A0A1S3HPH1 A0A195DG47 A0A182PUM4 A0A369RUA1 A0A195CJ24 A0A1B6GNG2 A0A2B4R799 A0A1S3ID49 A0A182G4E6 Q7PSL0 B3RXG7 A0A182YM72 A0A1L8DZF9 A0A182R867 A0A369RY28 A0A182SCF1 A0A1Q3FIY4 A0A2J7QLS2 A0A2P8Y7W7 A0A182WW22 A0A2C9KWG2 A0A1S3J1W1 A0A182QR12 A0A1L8DYZ5 A0A1S3J1C6 A0A182V899 A0A2J7QLS4 A0A182FB05 A0A182TRT7 A0A182TMS7 F5HJ76
A0A194PZH5 A0A194R0D8 A0A194PTD5 A0A212FII9 A0A194PT23 A0A194QZM0 I4DPZ6 A0A212FIH9 A0A194R162 A0A194PUB9 A0A0L7L8I0 A0A194PUC3 A0A194PT28 A0A2A4J3S6 A0A2A4JNM0 A0A212ETT6 A0A2H1V0T1 G8GE17 A0A0L7L784 A0A2H1VTP5 H9JXM0 A0A2A4J232 A0A2H1WNI5 A0A0L7LP26 A0A0L7L6L8 H9J2F6 A0A194QRL9 A0A194RLN8 A0A194PGU6 A0A2H1VKC8 A0A0L7L765 A0A2H1V0C4 A0A2H1WJN9 A0A194PHR8 A0A2A4JMM7 A0A2W1BTP3 A0A2P8YFR7 A0A212EHJ8 A0A194PPL0 H9J5Y1 A0A0L7LH10 A0A0N0BDE6 Q17Q46 A0A0L7R1M1 A0A1S4EV05 A0A088A0E6 A0A2A3EQ70 A0A154P2Q1 K7J1G6 A0A195FCB7 A0A2C9GG62 A0A3M1BMG2 A0A158NKN5 A0A0M8K9R9 A0A0P6YM72 J9HHI9 A0A232FI37 A0A151WRK1 B3RJA0 A0A1B6G7E7 A0A1S3HPH1 A0A195DG47 A0A182PUM4 A0A369RUA1 A0A195CJ24 A0A1B6GNG2 A0A2B4R799 A0A1S3ID49 A0A182G4E6 Q7PSL0 B3RXG7 A0A182YM72 A0A1L8DZF9 A0A182R867 A0A369RY28 A0A182SCF1 A0A1Q3FIY4 A0A2J7QLS2 A0A2P8Y7W7 A0A182WW22 A0A2C9KWG2 A0A1S3J1W1 A0A182QR12 A0A1L8DYZ5 A0A1S3J1C6 A0A182V899 A0A2J7QLS4 A0A182FB05 A0A182TRT7 A0A182TMS7 F5HJ76
PDB
3NI2
E-value=1.12181e-14,
Score=191
Ontologies
GO
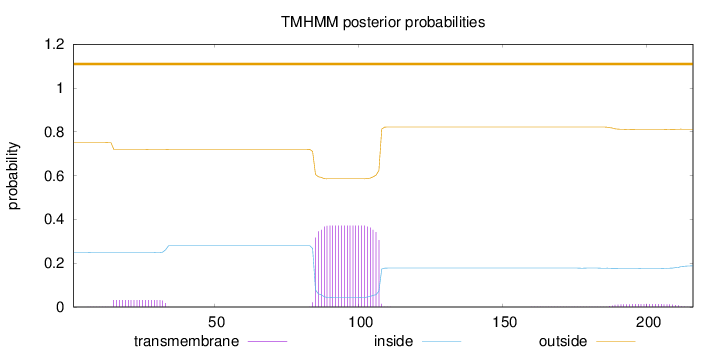
Topology
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.24381
Exp number, first 60 AAs:
0.59449
Total prob of N-in:
0.24875
outside
1 - 216
Population Genetic Test Statistics
Pi
227.199412
Theta
158.399162
Tajima's D
1.801236
CLR
0.101986
CSRT
0.849357532123394
Interpretation
Uncertain
