Gene
KWMTBOMO00844
Pre Gene Modal
BGIBMGA009681
Annotation
PREDICTED:_uncharacterized_protein_LOC106129482_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.318
Sequence
CDS
ATGAGGTGTTTATTGATAATAAGCGCGCTGCTCGCGACCGCGCGTTCCAAGCAAATCCCGCTAACAGATTTACCGGGAACGACGGTAGCTGCGGTATCCGAGTCAGATACCACGTCGTGGCCCGGCGCCGTCGCTGCCCTGGACTCTCTCACCGTCGCCTGCGAGAAAGACAGCATGCAAGTCACCATTACACTGGCGAAACGGGACCCCGAAATCAATAGCATCTACGACATATTCAATGGGATCGTCTACCCTGCTGGACTTGGGAGCAATTCGAGCTGTCTCCGAGAATACGTGGCAGAGCGGGGAGACCTACAGTACACCGTGCCTTTGAAGGGCTGCAATACCATGTCCACTGATAACGACGACGGCACAGTTGAGTACTACAACAACATCATAGTTCAGCCGCACTTGAAGCTGGTGACCGGCCAGGGTCGAGGCTACCACGTGAGGTGCAGGTACCGGCGCCGCGACCTCACACATTTCCACGTGTACCGACCGAGGGCCCACGCCGACAGACTCACCAATTCACTATCCGATGATTTTAATGACAACGAGGAGTTTGACGAGGATTCAGGATTGTTGCCGTCTGCCACGATGAAGATATATAAGGGTGACCCTAAAAATAAAGAGGAGGCATCAAACGTGAAGATCGGCGACACATTGACCCTGGTCGTGGCGCTGGAGAAACAGCGGCGGTTCGGTTTGCTGGTCTCCGAGTGTTCTGTTAAGGATGGACTGGGCTGGTCGGAGCAGACCCTCATCGCTGATAATGGATGTCCATTAGACGCTGAGATAATGGGCATGTTCCAATACTCGAGCGACAAGCAAGAGGCGCATGTGGCGTACCCCGCGCATAAGTTCCCGTACACAGCCAGCGTGTACTACACGTGTGCCGTCAAGTTGTGCGACCTGCAGCACCCGACGGACTGCGAACCTTGCAAGATAAAGGAAAGGAGCCGCCGCGACACAGACGACGAAGGCTCCCCAGCGACAGTGGAACTCTTCTCCGGCTTATATGTCAACGAGCCAGATTCGCTTGACAACGACGACGTTATCAGCGAAAAGAAAGAGGACGAGATATGTATTTCGCAACGTAATTTCGCCATCGGCATATGCATAGCCGGCGTTATTCTGATGGTGTGCGTCATAGCGGCTATCGCCTTCATACTGGCTCGACGAAGGAATCCGAAGGCGTACTCGCGGACTGGCAGCTCCCTGTACAGCGGCCCTTACACTAACACCGGCTATTCACACACAAGCTAA
Protein
MRCLLIISALLATARSKQIPLTDLPGTTVAAVSESDTTSWPGAVAALDSLTVACEKDSMQVTITLAKRDPEINSIYDIFNGIVYPAGLGSNSSCLREYVAERGDLQYTVPLKGCNTMSTDNDDGTVEYYNNIIVQPHLKLVTGQGRGYHVRCRYRRRDLTHFHVYRPRAHADRLTNSLSDDFNDNEEFDEDSGLLPSATMKIYKGDPKNKEEASNVKIGDTLTLVVALEKQRRFGLLVSECSVKDGLGWSEQTLIADNGCPLDAEIMGMFQYSSDKQEAHVAYPAHKFPYTASVYYTCAVKLCDLQHPTDCEPCKIKERSRRDTDDEGSPATVELFSGLYVNEPDSLDNDDVISEKKEDEICISQRNFAIGICIAGVILMVCVIAAIAFILARRRNPKAYSRTGSSLYSGPYTNTGYSHTS
Summary
Uniprot
A0A2A4JRZ3
A0A194R0D3
A0A194PUB4
H9JJI0
A0A2H1VKM8
A0A2H1W3P4
+ More
D6WRI4 A0A1Y1MK04 A0A1Y1MG57 A0A2J7Q1R9 A0A067QQV6 A0A3L8DEV3 A0A0M9A8R5 A0A154P1K1 A0A0C9QIY3 A0A0C9RZ56 A0A310S839 A0A195DFI8 K7IRX9 A0A084WJU9 A0A0K8TK78 A0A1B6HCP0 A0A1B6KC75 A0A1B6MRD1 E0VID3 A0A182L393 Q7QIT1 A0A182II10 A0A0P6IY40 A0A182NZ03 A0A1B6CA67 A0A1B0GP52 A0A1L8DA00 A0A023ERW3 A0A2A3E997 A0A1Q3FZM4 A0A151XH81 A0A1B6DUH5 B4KQ57 A0A0Q9X813 A0A0P6ATZ6 A0A0Q9XI05 A0A164PAQ9 A0A088AMC4 A0A0N7ZPS0 A0A2M3ZEE5 A0A2M3Z039 A0A0P6IAM8 A0A0P6A4G2 E2B1Q3 A0A0J7KMR4 A0A0P4X3R6 A0A2M4A232 A0A2M4DLY2 A0A2M4DM15 A0A1J1IVA0 J9JLL3 A0A2S2NRF2 A0A0M5IZ38 A0A023ETS9 A0A2H8TXU2 A0A2S2Q2R3 A0A2M4BNB0 B4PBD4 B4LJI5 Q8IRK0 A0A195CWW2 B3NQU9 A0A195BPP2 A0A0J9UAH8 A0A0P5X1J6 A0A1W4WBH4 B3MG88 A0A0P5ADN8 E9H6X2 B5E198 B4H563 N6TT58 U5EQP5 A0A0P5UCP0 A0A232F6F1 B4MP02 A0A224XRQ8 A0A195EYM0 B4IH44 A0A087TJD0 A0A023FNC2 U4UB72 A0A023G444 A0A224Z4A9 A0A131YG85 R4G3D2 A0A147BR43 A0A1E1X721 A0A0A9VXF5 A0A182M7H5 A0A182J9I6 A0A2P8XG86
D6WRI4 A0A1Y1MK04 A0A1Y1MG57 A0A2J7Q1R9 A0A067QQV6 A0A3L8DEV3 A0A0M9A8R5 A0A154P1K1 A0A0C9QIY3 A0A0C9RZ56 A0A310S839 A0A195DFI8 K7IRX9 A0A084WJU9 A0A0K8TK78 A0A1B6HCP0 A0A1B6KC75 A0A1B6MRD1 E0VID3 A0A182L393 Q7QIT1 A0A182II10 A0A0P6IY40 A0A182NZ03 A0A1B6CA67 A0A1B0GP52 A0A1L8DA00 A0A023ERW3 A0A2A3E997 A0A1Q3FZM4 A0A151XH81 A0A1B6DUH5 B4KQ57 A0A0Q9X813 A0A0P6ATZ6 A0A0Q9XI05 A0A164PAQ9 A0A088AMC4 A0A0N7ZPS0 A0A2M3ZEE5 A0A2M3Z039 A0A0P6IAM8 A0A0P6A4G2 E2B1Q3 A0A0J7KMR4 A0A0P4X3R6 A0A2M4A232 A0A2M4DLY2 A0A2M4DM15 A0A1J1IVA0 J9JLL3 A0A2S2NRF2 A0A0M5IZ38 A0A023ETS9 A0A2H8TXU2 A0A2S2Q2R3 A0A2M4BNB0 B4PBD4 B4LJI5 Q8IRK0 A0A195CWW2 B3NQU9 A0A195BPP2 A0A0J9UAH8 A0A0P5X1J6 A0A1W4WBH4 B3MG88 A0A0P5ADN8 E9H6X2 B5E198 B4H563 N6TT58 U5EQP5 A0A0P5UCP0 A0A232F6F1 B4MP02 A0A224XRQ8 A0A195EYM0 B4IH44 A0A087TJD0 A0A023FNC2 U4UB72 A0A023G444 A0A224Z4A9 A0A131YG85 R4G3D2 A0A147BR43 A0A1E1X721 A0A0A9VXF5 A0A182M7H5 A0A182J9I6 A0A2P8XG86
Pubmed
26354079
19121390
18362917
19820115
28004739
24845553
+ More
30249741 20075255 24438588 26369729 20566863 20966253 12364791 14747013 17210077 26999592 24945155 26483478 17994087 20798317 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 21292972 15632085 23185243 23537049 28648823 28797301 26830274 29652888 28503490 25401762 26823975 29403074
30249741 20075255 24438588 26369729 20566863 20966253 12364791 14747013 17210077 26999592 24945155 26483478 17994087 20798317 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 21292972 15632085 23185243 23537049 28648823 28797301 26830274 29652888 28503490 25401762 26823975 29403074
EMBL
NWSH01000684
PCG74805.1
KQ460890
KPJ10984.1
KQ459593
KPI96563.1
+ More
BABH01017559 ODYU01003055 SOQ41351.1 ODYU01006129 SOQ47710.1 KQ971351 EFA06573.1 GEZM01032293 JAV84685.1 GEZM01032294 JAV84684.1 NEVH01019380 PNF22525.1 KK853496 KDR07206.1 QOIP01000009 RLU18924.1 KQ435711 KOX79527.1 KQ434796 KZC05806.1 GBYB01014698 JAG84465.1 GBYB01014695 JAG84462.1 KQ764883 OAD54311.1 KQ980903 KYN11655.1 ATLV01024074 KE525348 KFB50493.1 GDAI01002926 JAI14677.1 GECU01035315 JAS72391.1 GEBQ01030936 JAT09041.1 GEBQ01001532 JAT38445.1 DS235196 EEB13139.1 AAAB01008807 EAA03968.4 APCN01001489 GDUN01000204 JAN95715.1 GEDC01026926 JAS10372.1 AJVK01033134 AJVK01033135 GFDF01010940 JAV03144.1 JXUM01002660 JXUM01002661 JXUM01002662 JXUM01002663 JXUM01002664 JXUM01002665 JXUM01002666 JXUM01002667 GAPW01001653 KQ560143 JAC11945.1 KXJ84198.1 KZ288340 PBC27759.1 GFDL01002062 JAV32983.1 KQ982130 KYQ59756.1 GEDC01007972 JAS29326.1 CH933808 EDW09185.2 KRG04511.1 GDIP01038354 JAM65361.1 KRG04510.1 LRGB01002648 KZS06663.1 GDIP01224577 JAI98824.1 GGFM01006100 MBW26851.1 GGFM01001141 MBW21892.1 GDIQ01014248 JAN80489.1 GDIP01039550 JAM64165.1 GL444953 EFN60409.1 LBMM01005354 KMQ91582.1 GDIP01250811 JAI72590.1 GGFK01001553 MBW34874.1 GGFL01014386 MBW78564.1 GGFL01014387 MBW78565.1 CVRI01000059 CRL03070.1 ABLF02033007 GGMR01007184 MBY19803.1 CP012524 ALC40748.1 GAPW01001774 JAC11824.1 GFXV01006816 MBW18621.1 GGMS01002823 MBY72026.1 GGFJ01005343 MBW54484.1 CM000158 EDW92566.1 KRK00580.1 KRK00581.1 CH940648 EDW61553.1 KRF80059.1 AE013599 BT003795 BT011382 AY862157 AAM68320.2 AAO41478.1 AAR96174.1 AAW51949.1 ACZ94567.1 AHN56644.1 KQ977185 KYN05042.1 CH954179 EDV57032.1 KQS63135.1 KQ976432 KYM87462.1 CM002911 KMY96379.1 KMY96380.1 KMY96381.1 GDIP01079054 JAM24661.1 CH902619 EDV37791.1 KPU77035.1 GDIP01200583 JAJ22819.1 GL732599 EFX72491.1 CM000071 EDY69486.1 KRT03092.1 KRT03093.1 CH479210 EDW32899.1 APGK01019949 APGK01019950 APGK01019951 APGK01019952 KB740150 ENN81248.1 GANO01004137 JAB55734.1 GDIP01116396 JAL87318.1 NNAY01000899 OXU26019.1 CH963848 EDW73841.2 GFTR01005735 JAW10691.1 KQ981905 KYN33390.1 CH480838 EDW49220.1 KK115487 KFM65219.1 GBBK01001723 JAC22759.1 KB632173 ERL89573.1 GBBM01006432 JAC28986.1 GFPF01010635 MAA21781.1 GEDV01011065 JAP77492.1 ACPB03000430 ACPB03000431 GAHY01001786 JAA75724.1 GEGO01002170 JAR93234.1 GFAC01004337 JAT94851.1 GBHO01043733 GBRD01000595 GDHC01002599 JAF99870.1 JAG65226.1 JAQ16030.1 AXCM01002461 PYGN01002240 PSN31033.1
BABH01017559 ODYU01003055 SOQ41351.1 ODYU01006129 SOQ47710.1 KQ971351 EFA06573.1 GEZM01032293 JAV84685.1 GEZM01032294 JAV84684.1 NEVH01019380 PNF22525.1 KK853496 KDR07206.1 QOIP01000009 RLU18924.1 KQ435711 KOX79527.1 KQ434796 KZC05806.1 GBYB01014698 JAG84465.1 GBYB01014695 JAG84462.1 KQ764883 OAD54311.1 KQ980903 KYN11655.1 ATLV01024074 KE525348 KFB50493.1 GDAI01002926 JAI14677.1 GECU01035315 JAS72391.1 GEBQ01030936 JAT09041.1 GEBQ01001532 JAT38445.1 DS235196 EEB13139.1 AAAB01008807 EAA03968.4 APCN01001489 GDUN01000204 JAN95715.1 GEDC01026926 JAS10372.1 AJVK01033134 AJVK01033135 GFDF01010940 JAV03144.1 JXUM01002660 JXUM01002661 JXUM01002662 JXUM01002663 JXUM01002664 JXUM01002665 JXUM01002666 JXUM01002667 GAPW01001653 KQ560143 JAC11945.1 KXJ84198.1 KZ288340 PBC27759.1 GFDL01002062 JAV32983.1 KQ982130 KYQ59756.1 GEDC01007972 JAS29326.1 CH933808 EDW09185.2 KRG04511.1 GDIP01038354 JAM65361.1 KRG04510.1 LRGB01002648 KZS06663.1 GDIP01224577 JAI98824.1 GGFM01006100 MBW26851.1 GGFM01001141 MBW21892.1 GDIQ01014248 JAN80489.1 GDIP01039550 JAM64165.1 GL444953 EFN60409.1 LBMM01005354 KMQ91582.1 GDIP01250811 JAI72590.1 GGFK01001553 MBW34874.1 GGFL01014386 MBW78564.1 GGFL01014387 MBW78565.1 CVRI01000059 CRL03070.1 ABLF02033007 GGMR01007184 MBY19803.1 CP012524 ALC40748.1 GAPW01001774 JAC11824.1 GFXV01006816 MBW18621.1 GGMS01002823 MBY72026.1 GGFJ01005343 MBW54484.1 CM000158 EDW92566.1 KRK00580.1 KRK00581.1 CH940648 EDW61553.1 KRF80059.1 AE013599 BT003795 BT011382 AY862157 AAM68320.2 AAO41478.1 AAR96174.1 AAW51949.1 ACZ94567.1 AHN56644.1 KQ977185 KYN05042.1 CH954179 EDV57032.1 KQS63135.1 KQ976432 KYM87462.1 CM002911 KMY96379.1 KMY96380.1 KMY96381.1 GDIP01079054 JAM24661.1 CH902619 EDV37791.1 KPU77035.1 GDIP01200583 JAJ22819.1 GL732599 EFX72491.1 CM000071 EDY69486.1 KRT03092.1 KRT03093.1 CH479210 EDW32899.1 APGK01019949 APGK01019950 APGK01019951 APGK01019952 KB740150 ENN81248.1 GANO01004137 JAB55734.1 GDIP01116396 JAL87318.1 NNAY01000899 OXU26019.1 CH963848 EDW73841.2 GFTR01005735 JAW10691.1 KQ981905 KYN33390.1 CH480838 EDW49220.1 KK115487 KFM65219.1 GBBK01001723 JAC22759.1 KB632173 ERL89573.1 GBBM01006432 JAC28986.1 GFPF01010635 MAA21781.1 GEDV01011065 JAP77492.1 ACPB03000430 ACPB03000431 GAHY01001786 JAA75724.1 GEGO01002170 JAR93234.1 GFAC01004337 JAT94851.1 GBHO01043733 GBRD01000595 GDHC01002599 JAF99870.1 JAG65226.1 JAQ16030.1 AXCM01002461 PYGN01002240 PSN31033.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000005204
UP000007266
UP000235965
+ More
UP000027135 UP000279307 UP000053105 UP000076502 UP000078492 UP000002358 UP000030765 UP000009046 UP000075882 UP000007062 UP000075840 UP000075884 UP000092462 UP000069940 UP000249989 UP000242457 UP000075809 UP000009192 UP000076858 UP000005203 UP000000311 UP000036403 UP000183832 UP000007819 UP000092553 UP000002282 UP000008792 UP000000803 UP000078542 UP000008711 UP000078540 UP000192221 UP000007801 UP000000305 UP000001819 UP000008744 UP000019118 UP000215335 UP000007798 UP000078541 UP000001292 UP000054359 UP000030742 UP000015103 UP000075883 UP000075880 UP000245037
UP000027135 UP000279307 UP000053105 UP000076502 UP000078492 UP000002358 UP000030765 UP000009046 UP000075882 UP000007062 UP000075840 UP000075884 UP000092462 UP000069940 UP000249989 UP000242457 UP000075809 UP000009192 UP000076858 UP000005203 UP000000311 UP000036403 UP000183832 UP000007819 UP000092553 UP000002282 UP000008792 UP000000803 UP000078542 UP000008711 UP000078540 UP000192221 UP000007801 UP000000305 UP000001819 UP000008744 UP000019118 UP000215335 UP000007798 UP000078541 UP000001292 UP000054359 UP000030742 UP000015103 UP000075883 UP000075880 UP000245037
Pfam
PF00100 Zona_pellucida
Interpro
IPR001507
ZP_dom
ProteinModelPortal
A0A2A4JRZ3
A0A194R0D3
A0A194PUB4
H9JJI0
A0A2H1VKM8
A0A2H1W3P4
+ More
D6WRI4 A0A1Y1MK04 A0A1Y1MG57 A0A2J7Q1R9 A0A067QQV6 A0A3L8DEV3 A0A0M9A8R5 A0A154P1K1 A0A0C9QIY3 A0A0C9RZ56 A0A310S839 A0A195DFI8 K7IRX9 A0A084WJU9 A0A0K8TK78 A0A1B6HCP0 A0A1B6KC75 A0A1B6MRD1 E0VID3 A0A182L393 Q7QIT1 A0A182II10 A0A0P6IY40 A0A182NZ03 A0A1B6CA67 A0A1B0GP52 A0A1L8DA00 A0A023ERW3 A0A2A3E997 A0A1Q3FZM4 A0A151XH81 A0A1B6DUH5 B4KQ57 A0A0Q9X813 A0A0P6ATZ6 A0A0Q9XI05 A0A164PAQ9 A0A088AMC4 A0A0N7ZPS0 A0A2M3ZEE5 A0A2M3Z039 A0A0P6IAM8 A0A0P6A4G2 E2B1Q3 A0A0J7KMR4 A0A0P4X3R6 A0A2M4A232 A0A2M4DLY2 A0A2M4DM15 A0A1J1IVA0 J9JLL3 A0A2S2NRF2 A0A0M5IZ38 A0A023ETS9 A0A2H8TXU2 A0A2S2Q2R3 A0A2M4BNB0 B4PBD4 B4LJI5 Q8IRK0 A0A195CWW2 B3NQU9 A0A195BPP2 A0A0J9UAH8 A0A0P5X1J6 A0A1W4WBH4 B3MG88 A0A0P5ADN8 E9H6X2 B5E198 B4H563 N6TT58 U5EQP5 A0A0P5UCP0 A0A232F6F1 B4MP02 A0A224XRQ8 A0A195EYM0 B4IH44 A0A087TJD0 A0A023FNC2 U4UB72 A0A023G444 A0A224Z4A9 A0A131YG85 R4G3D2 A0A147BR43 A0A1E1X721 A0A0A9VXF5 A0A182M7H5 A0A182J9I6 A0A2P8XG86
D6WRI4 A0A1Y1MK04 A0A1Y1MG57 A0A2J7Q1R9 A0A067QQV6 A0A3L8DEV3 A0A0M9A8R5 A0A154P1K1 A0A0C9QIY3 A0A0C9RZ56 A0A310S839 A0A195DFI8 K7IRX9 A0A084WJU9 A0A0K8TK78 A0A1B6HCP0 A0A1B6KC75 A0A1B6MRD1 E0VID3 A0A182L393 Q7QIT1 A0A182II10 A0A0P6IY40 A0A182NZ03 A0A1B6CA67 A0A1B0GP52 A0A1L8DA00 A0A023ERW3 A0A2A3E997 A0A1Q3FZM4 A0A151XH81 A0A1B6DUH5 B4KQ57 A0A0Q9X813 A0A0P6ATZ6 A0A0Q9XI05 A0A164PAQ9 A0A088AMC4 A0A0N7ZPS0 A0A2M3ZEE5 A0A2M3Z039 A0A0P6IAM8 A0A0P6A4G2 E2B1Q3 A0A0J7KMR4 A0A0P4X3R6 A0A2M4A232 A0A2M4DLY2 A0A2M4DM15 A0A1J1IVA0 J9JLL3 A0A2S2NRF2 A0A0M5IZ38 A0A023ETS9 A0A2H8TXU2 A0A2S2Q2R3 A0A2M4BNB0 B4PBD4 B4LJI5 Q8IRK0 A0A195CWW2 B3NQU9 A0A195BPP2 A0A0J9UAH8 A0A0P5X1J6 A0A1W4WBH4 B3MG88 A0A0P5ADN8 E9H6X2 B5E198 B4H563 N6TT58 U5EQP5 A0A0P5UCP0 A0A232F6F1 B4MP02 A0A224XRQ8 A0A195EYM0 B4IH44 A0A087TJD0 A0A023FNC2 U4UB72 A0A023G444 A0A224Z4A9 A0A131YG85 R4G3D2 A0A147BR43 A0A1E1X721 A0A0A9VXF5 A0A182M7H5 A0A182J9I6 A0A2P8XG86
Ontologies
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.996508
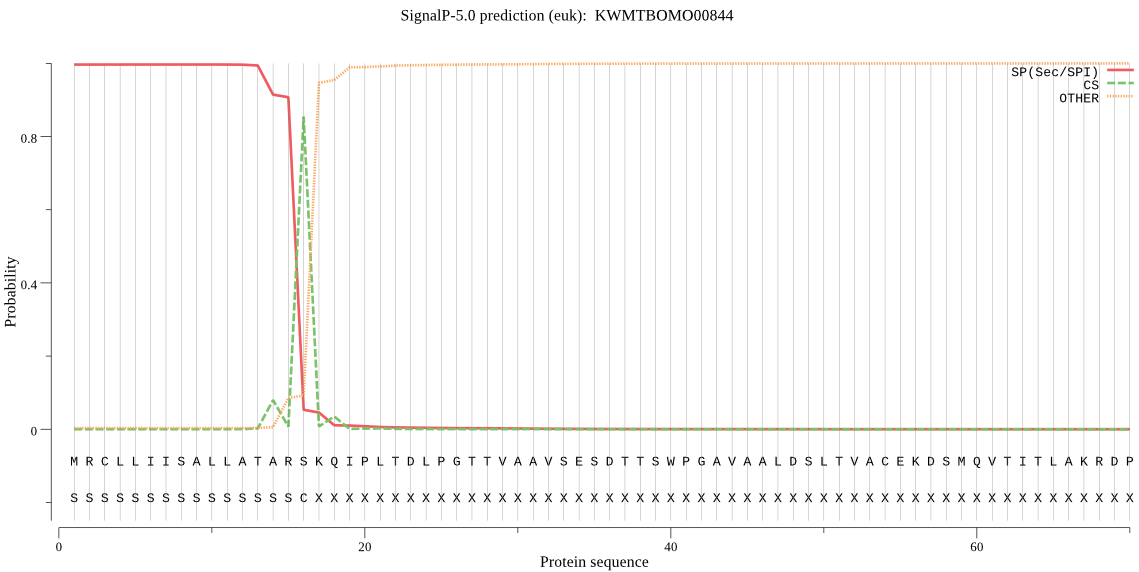
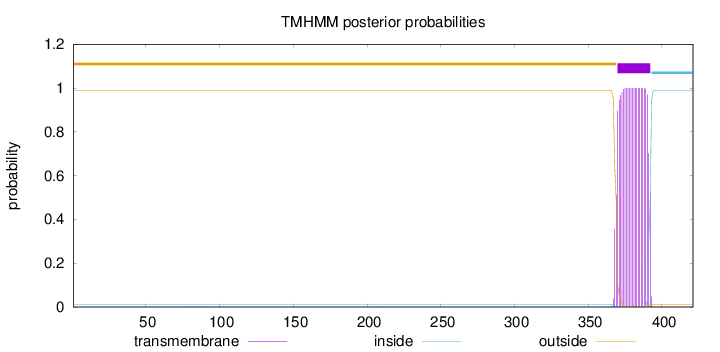
Length:
421
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.94376
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.01116
outside
1 - 369
TMhelix
370 - 392
inside
393 - 421
Population Genetic Test Statistics
Pi
23.353553
Theta
152.04326
Tajima's D
1.500285
CLR
0.044833
CSRT
0.789810509474526
Interpretation
Uncertain
