Gene
KWMTBOMO00842
Pre Gene Modal
BGIBMGA009755
Annotation
PREDICTED:_protein_lunapark-B_[Papilio_machaon]
Full name
Endoplasmic reticulum junction formation protein lunapark-B
Alternative Name
ER junction formation factor lunapark
Location in the cell
Nuclear Reliability : 2.665
Sequence
CDS
ATGGGCCTTATAATTTCAAGATTTCGCCGCAAGAAAACAACAACAGAAATACTCGAACGACTAGAAAACGATATAAAAAGCATAGAAAGAGATGGCCAATACAAGGAGCAAACCCACAAACGCGTTATAGGATACGTTATGGCGTATTCAGTGGGTCTATACGTGTTATTTGCGGTCCTTTACTACTACATGTACGTGGGGAGGAGTCAGTATTGGTTGCATTCGTTGCTGTATGCCTCGCCTTTACTGGTTTTGCCCGTGCTTGTGATATTCTTACGGTCATCAATATCCTGGTATTATAATTGGATGATAAACAAGAACAGGATGAAGTTAAGCAAAATGAGGGAGGATAAAAAGAAGATATTGGAAGAGGTGATGAACACAGAGACATATAAGGTTGCCAAAGAAATCCTTGACAAATACGGAAGTCCAGACGACCAGTCGAAATCGTTGAAGCCGTTCGTACCAACCCTGAACGTACCCGGTAATCTTCCTAGTTCACCCCCAGCAACACCGGGACAGTTAAGGCAGAGACAGCTAGCGTTGCAGACTTCGACGCCCATACCGAATAAAATGAACATGTTGGCTATAAACAACACGCCGAATGCTCTGTACAAAGGACCGCGTCTTAGATCGGACCAGTTGCCCAGGCCACTGCTCGACAAGGACCGGTCTGCTCTCGACAAAGTGGTGGACTATCTTCTCAAGGACGGACCGAATCATCGGATGGCGCTGATATGCAGGGAATGCTTCTCGCACAACGGCATGGCGATGGCCGAAGAGTACGAGTACGTGTCGTACGTGTGCGCGTACTGCGGGCGTCTCAACCCGGCGCGGAAGCAGAGGCCGGCCGCGCCGCTGCTGAGCCCGAAGGCGCTGCCGCCGGCCGTAAGGAGGATGCACGGTGGAAGTGACGATTCTTCGGTAGCCAGTTCCGGAAGTGAAAGCGAAGACGAGAAGAATGGAGGAAATCATTGTAATAATTTTGAGAAATCGATGAGCGAGTCGGATCGTCCCCCCTCCCCGGTTGAGGCGGACCCGAAATCCGACGAAGAAAAAAAAGAAGACTGA
Protein
MGLIISRFRRKKTTTEILERLENDIKSIERDGQYKEQTHKRVIGYVMAYSVGLYVLFAVLYYYMYVGRSQYWLHSLLYASPLLVLPVLVIFLRSSISWYYNWMINKNRMKLSKMREDKKKILEEVMNTETYKVAKEILDKYGSPDDQSKSLKPFVPTLNVPGNLPSSPPATPGQLRQRQLALQTSTPIPNKMNMLAINNTPNALYKGPRLRSDQLPRPLLDKDRSALDKVVDYLLKDGPNHRMALICRECFSHNGMAMAEEYEYVSYVCAYCGRLNPARKQRPAAPLLSPKALPPAVRRMHGGSDDSSVASSGSESEDEKNGGNHCNNFEKSMSESDRPPSPVEADPKSDEEKKED
Summary
Description
Endoplasmic reticulum (ER)-shaping membrane protein that plays a role in determining ER morphology. Involved in the stabilization of nascent three-way ER tubular junctions within the ER network. May also play a role as a curvature-stabilizing protein within three-way ER tubular junction network (By similarity).
Subunit
Homodimer; homodimerization requires the C4-type zinc finger motif and decreases during mitosis in a phosphorylation-dependent manner.
Similarity
Belongs to the lunapark family.
Keywords
Coiled coil
Complete proteome
Endoplasmic reticulum
Membrane
Metal-binding
Reference proteome
Transmembrane
Transmembrane helix
Zinc
Zinc-finger
Feature
chain Endoplasmic reticulum junction formation protein lunapark-B
Uniprot
A0A194QZV1
A0A194PT16
A0A212EIF6
E1ZW26
A0A158NSC4
A0A0J7NTR1
+ More
A0A151I2S0 F4W5K3 A0A026W7Y3 A0A0C9RAD4 A0A088AUC3 A0A151JQL9 K7IP88 A0A2J7Q6Q7 A0A195D3X0 A0A0L7RBQ8 A0A2M4ASJ5 A0A0T6BCI7 A0A1Y1MZT0 A0A2P8Z7C9 A0A067QZC6 U5EPS4 T1HY37 A0A2M3ZKM9 A0A1A9WYU8 A0A1B6BWI0 A0A2M3ZAW2 A0A2A3EMU7 A0A023EYR4 A0A2M4BND6 A0A310SLU3 A0A1I8M1X2 A0A034VPA0 A0A182G051 T1GJ40 A0A0K8T8R3 A0A0K8V1G2 A0A069DSM8 A0A224XLN1 D6WRW1 A0A0A9YZY6 A0A182LRW0 A0A182RJ89 V4B9A4 A0A1Q3F482 A0A0K8TQ24 A0A1Q3F469 A0A1Q3F3Q9 A0A1Q3F3J7 A0A1Q3F3H5 A0A1Q3F3M4 A0A3B1IET6 A0A3B1IK64 A0A1Q3F3F1 A0A1Q3F3I5 A0A3B1KDJ3 A0A0A1X2G5 H2ZLJ1 W4ZIZ9 Q7Q3Q4 F1QDW0 A0A182W7F9 Q6PFM4 A0A232F464 A0A182WT26 A0A182L1X5 A0A0R4IYI7 W5JG27 A0A182HV23 A0A182Q1I5 A0A182VBE5 A0A1S4GXK9 B0XAQ3 A0A1A9XDG8 A0A336LNE8 A0A1B0BNG4 A0A1S4K935 A0A0P4WMA9 A0A0N7ZLN7 A0A0N7ZQ27 A0A0P6GQ79 A0A0P6DJE7 Q17D88 A0A0P6DM87 A0A0P6DY44 A0A182NTL3 F6WQS5 A0A1B0AFV8 A0A0P5ESW4 A0A0P5DTN3 A0A0P5XQR1 A0A0N8CDE9 A0A0P5YC81 A0A0P4YQQ0 A0A0P6EWN3 A0A0N7ZW88 A0A0N8CIQ7 A0A0N8CMN2
A0A151I2S0 F4W5K3 A0A026W7Y3 A0A0C9RAD4 A0A088AUC3 A0A151JQL9 K7IP88 A0A2J7Q6Q7 A0A195D3X0 A0A0L7RBQ8 A0A2M4ASJ5 A0A0T6BCI7 A0A1Y1MZT0 A0A2P8Z7C9 A0A067QZC6 U5EPS4 T1HY37 A0A2M3ZKM9 A0A1A9WYU8 A0A1B6BWI0 A0A2M3ZAW2 A0A2A3EMU7 A0A023EYR4 A0A2M4BND6 A0A310SLU3 A0A1I8M1X2 A0A034VPA0 A0A182G051 T1GJ40 A0A0K8T8R3 A0A0K8V1G2 A0A069DSM8 A0A224XLN1 D6WRW1 A0A0A9YZY6 A0A182LRW0 A0A182RJ89 V4B9A4 A0A1Q3F482 A0A0K8TQ24 A0A1Q3F469 A0A1Q3F3Q9 A0A1Q3F3J7 A0A1Q3F3H5 A0A1Q3F3M4 A0A3B1IET6 A0A3B1IK64 A0A1Q3F3F1 A0A1Q3F3I5 A0A3B1KDJ3 A0A0A1X2G5 H2ZLJ1 W4ZIZ9 Q7Q3Q4 F1QDW0 A0A182W7F9 Q6PFM4 A0A232F464 A0A182WT26 A0A182L1X5 A0A0R4IYI7 W5JG27 A0A182HV23 A0A182Q1I5 A0A182VBE5 A0A1S4GXK9 B0XAQ3 A0A1A9XDG8 A0A336LNE8 A0A1B0BNG4 A0A1S4K935 A0A0P4WMA9 A0A0N7ZLN7 A0A0N7ZQ27 A0A0P6GQ79 A0A0P6DJE7 Q17D88 A0A0P6DM87 A0A0P6DY44 A0A182NTL3 F6WQS5 A0A1B0AFV8 A0A0P5ESW4 A0A0P5DTN3 A0A0P5XQR1 A0A0N8CDE9 A0A0P5YC81 A0A0P4YQQ0 A0A0P6EWN3 A0A0N7ZW88 A0A0N8CIQ7 A0A0N8CMN2
Pubmed
EMBL
KQ460890
KPJ10982.1
KQ459593
KPI96561.1
AGBW02014667
OWR41251.1
+ More
GL434758 EFN74613.1 ADTU01024816 LBMM01001759 KMQ95805.1 KQ976543 KYM80985.1 GL887655 EGI70500.1 KK107348 QOIP01000010 EZA52212.1 RLU17934.1 GBYB01005220 JAG74987.1 KQ978642 KYN29499.1 NEVH01017460 PNF24266.1 KQ976881 KYN07583.1 KQ414617 KOC68357.1 GGFK01010430 MBW43751.1 LJIG01001931 KRT85009.1 GEZM01016728 GEZM01016727 JAV91164.1 PYGN01000163 PSN52408.1 KK853162 KDR10430.1 GANO01000066 JAB59805.1 ACPB03014864 GGFM01008325 MBW29076.1 GEDC01031657 JAS05641.1 GGFM01004880 MBW25631.1 KZ288205 PBC33133.1 GBBI01004399 JAC14313.1 GGFJ01005458 MBW54599.1 KQ763832 OAD54707.1 GAKP01015005 JAC43947.1 CAQQ02005852 CAQQ02005853 CAQQ02005854 GBRD01003880 GBRD01003879 JAG61942.1 GDHF01019874 JAI32440.1 GBGD01001791 JAC87098.1 GFTR01005698 JAW10728.1 KQ971351 EFA06588.1 GBHO01008509 GBHO01008508 GBHO01008506 GDHC01021193 JAG35095.1 JAG35096.1 JAG35098.1 JAP97435.1 AXCM01008268 KB203274 ESO85439.1 GFDL01012684 JAV22361.1 GDAI01001134 JAI16469.1 GFDL01012679 JAV22366.1 GFDL01012851 JAV22194.1 GFDL01012946 JAV22099.1 GFDL01012952 JAV22093.1 GFDL01012947 JAV22098.1 GFDL01012960 JAV22085.1 GFDL01012942 JAV22103.1 GBXI01009464 JAD04828.1 AAGJ04152145 AAAB01008964 EAA12393.2 BX546447 CT573297 BC057494 NNAY01001020 OXU25465.1 ADMH02001321 ETN63021.1 APCN01002491 AXCN02000795 DS232592 EDS43842.1 UFQT01000093 SSX19612.1 JXJN01017383 GDIP01254271 JAI69130.1 GDIP01233241 JAI90160.1 GDIP01223721 JAI99680.1 GDIQ01042623 JAN52114.1 GDIQ01085717 JAN09020.1 CH477298 EAT44341.1 GDIQ01090207 JAN04530.1 GDIQ01085718 GDIQ01085716 JAN09019.1 EAAA01000296 GDIP01143799 JAJ79603.1 GDIP01151398 JAJ72004.1 GDIP01068632 JAM35083.1 GDIP01142655 JAL61059.1 GDIP01059844 JAM43871.1 GDIP01225246 JAI98155.1 GDIQ01056966 JAN37771.1 GDIP01206433 JAJ16969.1 GDIP01127791 LRGB01000512 JAL75923.1 KZS18895.1 GDIP01116791 JAL86923.1
GL434758 EFN74613.1 ADTU01024816 LBMM01001759 KMQ95805.1 KQ976543 KYM80985.1 GL887655 EGI70500.1 KK107348 QOIP01000010 EZA52212.1 RLU17934.1 GBYB01005220 JAG74987.1 KQ978642 KYN29499.1 NEVH01017460 PNF24266.1 KQ976881 KYN07583.1 KQ414617 KOC68357.1 GGFK01010430 MBW43751.1 LJIG01001931 KRT85009.1 GEZM01016728 GEZM01016727 JAV91164.1 PYGN01000163 PSN52408.1 KK853162 KDR10430.1 GANO01000066 JAB59805.1 ACPB03014864 GGFM01008325 MBW29076.1 GEDC01031657 JAS05641.1 GGFM01004880 MBW25631.1 KZ288205 PBC33133.1 GBBI01004399 JAC14313.1 GGFJ01005458 MBW54599.1 KQ763832 OAD54707.1 GAKP01015005 JAC43947.1 CAQQ02005852 CAQQ02005853 CAQQ02005854 GBRD01003880 GBRD01003879 JAG61942.1 GDHF01019874 JAI32440.1 GBGD01001791 JAC87098.1 GFTR01005698 JAW10728.1 KQ971351 EFA06588.1 GBHO01008509 GBHO01008508 GBHO01008506 GDHC01021193 JAG35095.1 JAG35096.1 JAG35098.1 JAP97435.1 AXCM01008268 KB203274 ESO85439.1 GFDL01012684 JAV22361.1 GDAI01001134 JAI16469.1 GFDL01012679 JAV22366.1 GFDL01012851 JAV22194.1 GFDL01012946 JAV22099.1 GFDL01012952 JAV22093.1 GFDL01012947 JAV22098.1 GFDL01012960 JAV22085.1 GFDL01012942 JAV22103.1 GBXI01009464 JAD04828.1 AAGJ04152145 AAAB01008964 EAA12393.2 BX546447 CT573297 BC057494 NNAY01001020 OXU25465.1 ADMH02001321 ETN63021.1 APCN01002491 AXCN02000795 DS232592 EDS43842.1 UFQT01000093 SSX19612.1 JXJN01017383 GDIP01254271 JAI69130.1 GDIP01233241 JAI90160.1 GDIP01223721 JAI99680.1 GDIQ01042623 JAN52114.1 GDIQ01085717 JAN09020.1 CH477298 EAT44341.1 GDIQ01090207 JAN04530.1 GDIQ01085718 GDIQ01085716 JAN09019.1 EAAA01000296 GDIP01143799 JAJ79603.1 GDIP01151398 JAJ72004.1 GDIP01068632 JAM35083.1 GDIP01142655 JAL61059.1 GDIP01059844 JAM43871.1 GDIP01225246 JAI98155.1 GDIQ01056966 JAN37771.1 GDIP01206433 JAJ16969.1 GDIP01127791 LRGB01000512 JAL75923.1 KZS18895.1 GDIP01116791 JAL86923.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000000311
UP000005205
UP000036403
+ More
UP000078540 UP000007755 UP000053097 UP000279307 UP000005203 UP000078492 UP000002358 UP000235965 UP000078542 UP000053825 UP000245037 UP000027135 UP000015103 UP000091820 UP000242457 UP000095301 UP000069272 UP000015102 UP000007266 UP000075883 UP000075900 UP000030746 UP000018467 UP000007875 UP000007110 UP000007062 UP000000437 UP000075920 UP000215335 UP000076407 UP000075882 UP000000673 UP000075840 UP000075886 UP000075903 UP000002320 UP000092443 UP000092460 UP000008820 UP000075884 UP000008144 UP000092445 UP000076858
UP000078540 UP000007755 UP000053097 UP000279307 UP000005203 UP000078492 UP000002358 UP000235965 UP000078542 UP000053825 UP000245037 UP000027135 UP000015103 UP000091820 UP000242457 UP000095301 UP000069272 UP000015102 UP000007266 UP000075883 UP000075900 UP000030746 UP000018467 UP000007875 UP000007110 UP000007062 UP000000437 UP000075920 UP000215335 UP000076407 UP000075882 UP000000673 UP000075840 UP000075886 UP000075903 UP000002320 UP000092443 UP000092460 UP000008820 UP000075884 UP000008144 UP000092445 UP000076858
PRIDE
Pfam
Interpro
IPR040115
Lnp
+ More
IPR019273 Lunapark_dom
IPR022383 Lactate/malate_DH_C
IPR015955 Lactate_DH/Glyco_Ohase_4_C
IPR036291 NAD(P)-bd_dom_sf
IPR006463 MiaB_methiolase
IPR005839 Methylthiotransferase
IPR001236 Lactate/malate_DH_N
IPR006638 Elp3/MiaB/NifB
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR002792 TRAM_dom
IPR007197 rSAM
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
IPR019273 Lunapark_dom
IPR022383 Lactate/malate_DH_C
IPR015955 Lactate_DH/Glyco_Ohase_4_C
IPR036291 NAD(P)-bd_dom_sf
IPR006463 MiaB_methiolase
IPR005839 Methylthiotransferase
IPR001236 Lactate/malate_DH_N
IPR006638 Elp3/MiaB/NifB
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR002792 TRAM_dom
IPR007197 rSAM
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
Gene 3D
ProteinModelPortal
A0A194QZV1
A0A194PT16
A0A212EIF6
E1ZW26
A0A158NSC4
A0A0J7NTR1
+ More
A0A151I2S0 F4W5K3 A0A026W7Y3 A0A0C9RAD4 A0A088AUC3 A0A151JQL9 K7IP88 A0A2J7Q6Q7 A0A195D3X0 A0A0L7RBQ8 A0A2M4ASJ5 A0A0T6BCI7 A0A1Y1MZT0 A0A2P8Z7C9 A0A067QZC6 U5EPS4 T1HY37 A0A2M3ZKM9 A0A1A9WYU8 A0A1B6BWI0 A0A2M3ZAW2 A0A2A3EMU7 A0A023EYR4 A0A2M4BND6 A0A310SLU3 A0A1I8M1X2 A0A034VPA0 A0A182G051 T1GJ40 A0A0K8T8R3 A0A0K8V1G2 A0A069DSM8 A0A224XLN1 D6WRW1 A0A0A9YZY6 A0A182LRW0 A0A182RJ89 V4B9A4 A0A1Q3F482 A0A0K8TQ24 A0A1Q3F469 A0A1Q3F3Q9 A0A1Q3F3J7 A0A1Q3F3H5 A0A1Q3F3M4 A0A3B1IET6 A0A3B1IK64 A0A1Q3F3F1 A0A1Q3F3I5 A0A3B1KDJ3 A0A0A1X2G5 H2ZLJ1 W4ZIZ9 Q7Q3Q4 F1QDW0 A0A182W7F9 Q6PFM4 A0A232F464 A0A182WT26 A0A182L1X5 A0A0R4IYI7 W5JG27 A0A182HV23 A0A182Q1I5 A0A182VBE5 A0A1S4GXK9 B0XAQ3 A0A1A9XDG8 A0A336LNE8 A0A1B0BNG4 A0A1S4K935 A0A0P4WMA9 A0A0N7ZLN7 A0A0N7ZQ27 A0A0P6GQ79 A0A0P6DJE7 Q17D88 A0A0P6DM87 A0A0P6DY44 A0A182NTL3 F6WQS5 A0A1B0AFV8 A0A0P5ESW4 A0A0P5DTN3 A0A0P5XQR1 A0A0N8CDE9 A0A0P5YC81 A0A0P4YQQ0 A0A0P6EWN3 A0A0N7ZW88 A0A0N8CIQ7 A0A0N8CMN2
A0A151I2S0 F4W5K3 A0A026W7Y3 A0A0C9RAD4 A0A088AUC3 A0A151JQL9 K7IP88 A0A2J7Q6Q7 A0A195D3X0 A0A0L7RBQ8 A0A2M4ASJ5 A0A0T6BCI7 A0A1Y1MZT0 A0A2P8Z7C9 A0A067QZC6 U5EPS4 T1HY37 A0A2M3ZKM9 A0A1A9WYU8 A0A1B6BWI0 A0A2M3ZAW2 A0A2A3EMU7 A0A023EYR4 A0A2M4BND6 A0A310SLU3 A0A1I8M1X2 A0A034VPA0 A0A182G051 T1GJ40 A0A0K8T8R3 A0A0K8V1G2 A0A069DSM8 A0A224XLN1 D6WRW1 A0A0A9YZY6 A0A182LRW0 A0A182RJ89 V4B9A4 A0A1Q3F482 A0A0K8TQ24 A0A1Q3F469 A0A1Q3F3Q9 A0A1Q3F3J7 A0A1Q3F3H5 A0A1Q3F3M4 A0A3B1IET6 A0A3B1IK64 A0A1Q3F3F1 A0A1Q3F3I5 A0A3B1KDJ3 A0A0A1X2G5 H2ZLJ1 W4ZIZ9 Q7Q3Q4 F1QDW0 A0A182W7F9 Q6PFM4 A0A232F464 A0A182WT26 A0A182L1X5 A0A0R4IYI7 W5JG27 A0A182HV23 A0A182Q1I5 A0A182VBE5 A0A1S4GXK9 B0XAQ3 A0A1A9XDG8 A0A336LNE8 A0A1B0BNG4 A0A1S4K935 A0A0P4WMA9 A0A0N7ZLN7 A0A0N7ZQ27 A0A0P6GQ79 A0A0P6DJE7 Q17D88 A0A0P6DM87 A0A0P6DY44 A0A182NTL3 F6WQS5 A0A1B0AFV8 A0A0P5ESW4 A0A0P5DTN3 A0A0P5XQR1 A0A0N8CDE9 A0A0P5YC81 A0A0P4YQQ0 A0A0P6EWN3 A0A0N7ZW88 A0A0N8CIQ7 A0A0N8CMN2
Ontologies
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
Localizes at endoplasmic reticulum (ER) three-way tubular junctions, which represent crossing-points at which the tubules build a polygonal network. With evidence from 3 publications.
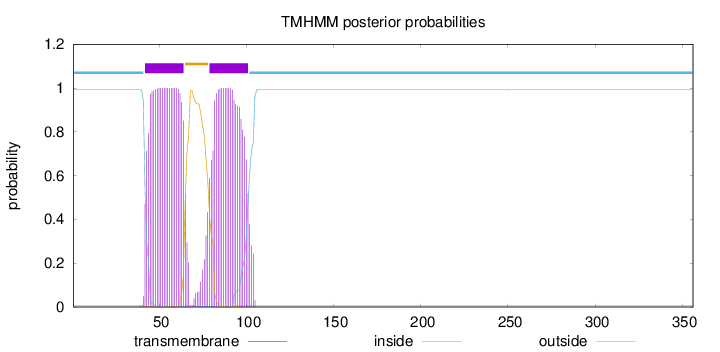
Length:
356
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.3959100000001
Exp number, first 60 AAs:
17.96154
Total prob of N-in:
0.99235
POSSIBLE N-term signal
sequence
inside
1 - 41
TMhelix
42 - 64
outside
65 - 78
TMhelix
79 - 101
inside
102 - 356
Population Genetic Test Statistics
Pi
229.65927
Theta
155.847956
Tajima's D
1.493094
CLR
0.024366
CSRT
0.787610619469027
Interpretation
Uncertain
