Gene
KWMTBOMO00839 Validated by peptides from experiments
Annotation
RNA_binding_motif_protein_Y14_[Bombyx_mori]
Full name
RNA-binding protein 8A
Alternative Name
Protein tsunagi
Location in the cell
Cytoplasmic Reliability : 1.839 Nuclear Reliability : 2.33
Sequence
CDS
ATGGCTGACGTGCTAGATATCGAAAATTCAGAAGAATTTGAAGTCGACGAAGATGGTGAACAGGGAATCGCTAGACTTAAGGAGAAGGCACGTAAGCGAAAAGGTCGTGGTTTTGGCAACGAAGCCGGTGGAGGCAGTGCTGCAGAACGGGGTAACCGCGGAAGATACGACAGTTTAGCTCCGGAAGGCGATAGCGGTACACCAGGACCTCAGAGATCAGTTGAAGGCTGGATCCTGTTTGTCAGTAATGTACATGAAGAAGCACAAGAAGAAGATATACAGAATCAATTTTCCGAGTTTGGTGAGATAAAGAATATTCATCTGAATCTGGACCGACGCACAGGCTTCCTCAAAGGCTACGCTCTAGTTGAGTACGAGACTTATAAACAAGCCGCTAGTGCACGTGAAGCATTGGATGGCGCAGATATACTCGGCCAAGCGATATCAGTTGATTGGTGCTTCGTCAAGGGACCCACTAAATCACATAAGAAGAGACGATAA
Protein
MADVLDIENSEEFEVDEDGEQGIARLKEKARKRKGRGFGNEAGGGSAAERGNRGRYDSLAPEGDSGTPGPQRSVEGWILFVSNVHEEAQEEDIQNQFSEFGEIKNIHLNLDRRTGFLKGYALVEYETYKQAASAREALDGADILGQAISVDWCFVKGPTKSHKKRR
Summary
Description
Core component of the splicing-dependent multiprotein exon junction complex (EJC) deposited at splice junctions on mRNAs.
Core component of the splicing-dependent multiprotein exon junction complex (EJC) deposited at splice junctions on mRNAs (PubMed:14973490, PubMed:24967911). Involved in exon definition of genes containing long introns, including the rolled/MAPK gene (PubMed:20946982, PubMed:20946983). The mago-tsu heterodimer interacts with the EJC key regulator Pym leading to EJC disassembly in the cytoplasm (PubMed:24967911). Has a role in oskar mRNA localization to the posterior pole of the developing oocyte (PubMed:11691839).
Core component of the splicing-dependent multiprotein exon junction complex (EJC) deposited at splice junctions on mRNAs (PubMed:14973490, PubMed:24967911). Involved in exon definition of genes containing long introns, including the rolled/MAPK gene (PubMed:20946982, PubMed:20946983). The mago-tsu heterodimer interacts with the EJC key regulator Pym leading to EJC disassembly in the cytoplasm (PubMed:24967911). Has a role in oskar mRNA localization to the posterior pole of the developing oocyte (PubMed:11691839).
Subunit
Heterodimer with MAGOH. Part of the mRNA splicing-dependent exon junction complex (EJC) complex; the core complex contains CASC3, EIF4A3, MAGOH and RBM8A.
Heterodimer (via RRM domain) with mago (PubMed:11691839, PubMed:12730685, PubMed:12704080). Part of the mRNA splicing-dependent exon junction complex (EJC) complex; the core complex contains btz/CASC3, eIF4AIII, mago and tsu/RBM8A (PubMed:14973490). Interacts with Pym (via N-terminus); the interaction is direct (PubMed:24967911, PubMed:14968132). Interacts with eIF4AIII (PubMed:14973490).
Heterodimer (via RRM domain) with mago (PubMed:11691839, PubMed:12730685, PubMed:12704080). Part of the mRNA splicing-dependent exon junction complex (EJC) complex; the core complex contains btz/CASC3, eIF4AIII, mago and tsu/RBM8A (PubMed:14973490). Interacts with Pym (via N-terminus); the interaction is direct (PubMed:24967911, PubMed:14968132). Interacts with eIF4AIII (PubMed:14973490).
Miscellaneous
'Tsunagi' means 'connection' or 'link' in Japanese.
Similarity
Belongs to the RBM8A family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Developmental protein
mRNA processing
mRNA splicing
mRNA transport
Nucleus
Reference proteome
RNA-binding
Transport
Feature
chain RNA-binding protein 8A
Uniprot
Q1HQB4
A0A194R0C8
A0A2A4J7A8
A0A2H1W6J9
G8A442
S4Q0B8
+ More
I4DL10 A0A1E1W6X4 A0A212F6A2 A0A1E1W5J3 A0A182GH26 A0A182QXI2 A0A182NJA3 A0A182MMH8 A0A1I8JUP7 A0A182PEQ9 Q175A5 A0A182YR35 A0A195E3N0 A0A158P1E8 A0A2M3ZB47 A0A2M4AQL4 A0A2M4C1L7 W5JH19 A0A182FCB2 A0A151XH13 A0A1Q3F892 A0A182UND7 A0A182K4Q3 A0A182UL70 A0A182WTS0 A0A182LL96 Q7Q5L6 A0A182HI17 E1ZWQ4 A0A026X1P1 A0A182J1W8 A0A151X7G2 A0A0J7KJJ2 F4X0Y4 A0A151I000 A0A195EYB5 A0A151IFT1 B0WKA7 K7J3K6 E2C0P8 R4V253 A0A2J7R3F3 A0A1B6J7C5 A0A0L7QRF4 U5EYB1 E2BCV8 A0A1B6G3G0 A0A1B6DSP1 A0A067QXR0 A0A0K8TRK4 A0A232F6P0 A0A1B0DR84 A0A1L8DHX2 A0A1B6LEK0 A0A154P2G6 A0A084WEJ1 A0A1L8EDI0 A0A0M8ZUV4 B4J9I4 A0A0M3QUF4 T1PH98 A0A1I8PAK5 W8AXF2 A0A0A1XID3 Q28ZB8 B3MDP6 B4ME32 A0A2P8YS07 B4KQJ1 A0A3B0JGF2 B4P3X1 B3N7L5 F2FB57 Q9V535 A0A1W4UZM2 B4GHZ2 A0A0K8VHU1 A0A034VVB5 B4NN37 A0A182WCC1 B4HSM1 B4QGR9 A0A0L0CKR0 A0A1A9W0Y8 A0A1A9UU10 A0A1A9ZUX9 D3TKL1 A0A1A9XY30 A0A1B0BKU7 A0A2R5L8F6 M9MSS7 A0A336KNV2 A0A2P2HY58 A0A1J1I4D2
I4DL10 A0A1E1W6X4 A0A212F6A2 A0A1E1W5J3 A0A182GH26 A0A182QXI2 A0A182NJA3 A0A182MMH8 A0A1I8JUP7 A0A182PEQ9 Q175A5 A0A182YR35 A0A195E3N0 A0A158P1E8 A0A2M3ZB47 A0A2M4AQL4 A0A2M4C1L7 W5JH19 A0A182FCB2 A0A151XH13 A0A1Q3F892 A0A182UND7 A0A182K4Q3 A0A182UL70 A0A182WTS0 A0A182LL96 Q7Q5L6 A0A182HI17 E1ZWQ4 A0A026X1P1 A0A182J1W8 A0A151X7G2 A0A0J7KJJ2 F4X0Y4 A0A151I000 A0A195EYB5 A0A151IFT1 B0WKA7 K7J3K6 E2C0P8 R4V253 A0A2J7R3F3 A0A1B6J7C5 A0A0L7QRF4 U5EYB1 E2BCV8 A0A1B6G3G0 A0A1B6DSP1 A0A067QXR0 A0A0K8TRK4 A0A232F6P0 A0A1B0DR84 A0A1L8DHX2 A0A1B6LEK0 A0A154P2G6 A0A084WEJ1 A0A1L8EDI0 A0A0M8ZUV4 B4J9I4 A0A0M3QUF4 T1PH98 A0A1I8PAK5 W8AXF2 A0A0A1XID3 Q28ZB8 B3MDP6 B4ME32 A0A2P8YS07 B4KQJ1 A0A3B0JGF2 B4P3X1 B3N7L5 F2FB57 Q9V535 A0A1W4UZM2 B4GHZ2 A0A0K8VHU1 A0A034VVB5 B4NN37 A0A182WCC1 B4HSM1 B4QGR9 A0A0L0CKR0 A0A1A9W0Y8 A0A1A9UU10 A0A1A9ZUX9 D3TKL1 A0A1A9XY30 A0A1B0BKU7 A0A2R5L8F6 M9MSS7 A0A336KNV2 A0A2P2HY58 A0A1J1I4D2
Pubmed
26354079
21193013
23622113
22651552
22118469
26483478
+ More
17510324 25244985 21347285 20920257 23761445 20966253 12364791 14747013 17210077 20798317 24508170 30249741 21719571 20075255 24845553 26369729 28648823 24438588 17994087 25315136 24495485 25830018 15632085 29403074 17550304 24416299 11691839 10731132 12537572 12537569 14973490 20946982 20946983 24967911 12730685 12704080 14968132 25348373 22936249 26108605 20353571
17510324 25244985 21347285 20920257 23761445 20966253 12364791 14747013 17210077 20798317 24508170 30249741 21719571 20075255 24845553 26369729 28648823 24438588 17994087 25315136 24495485 25830018 15632085 29403074 17550304 24416299 11691839 10731132 12537572 12537569 14973490 20946982 20946983 24967911 12730685 12704080 14968132 25348373 22936249 26108605 20353571
EMBL
DQ443138
DQ497206
FJ602787
ABF51227.1
ABF55971.1
ACM24353.1
+ More
KQ460890 KPJ10979.1 NWSH01002905 PCG67330.1 ODYU01006665 SOQ48710.1 GU108331 ADK56161.1 GAIX01000063 JAA92497.1 AK401978 KQ459593 BAM18600.1 KPI96557.1 GDQN01008331 GDQN01004536 GDQN01001903 JAT82723.1 JAT86518.1 JAT89151.1 AGBW02010065 OWR49266.1 GDQN01008797 GDQN01002866 JAT82257.1 JAT88188.1 JXUM01011038 KQ560322 KXJ83062.1 AXCN02000449 AXCM01001406 CH477404 EAT41626.1 KQ979709 KYN19479.1 ADTU01006397 GGFM01005016 MBW25767.1 GGFK01009749 MBW43070.1 GGFJ01010061 MBW59202.1 ADMH02001414 ETN62618.1 KQ982138 KYQ59605.1 GFDL01011273 JAV23772.1 AAAB01008960 EAA11513.3 APCN01005863 GL434886 EFN74382.1 KK107024 QOIP01000004 EZA62220.1 RLU23391.1 KQ982450 KYQ56264.1 LBMM01006642 KMQ90437.1 GL888505 EGI59886.1 KQ976633 KYM78876.1 KQ981928 KYN32879.1 KQ977771 KYN00006.1 DS231969 EDS29698.1 GL451846 EFN78442.1 KC740978 AGM32802.1 NEVH01007821 PNF35367.1 GECU01012636 JAS95070.1 KQ414784 KOC61081.1 GANO01000471 JAB59400.1 GL447410 EFN86470.1 GECZ01012805 JAS56964.1 GEDC01008649 JAS28649.1 KK852897 KDR14174.1 GDAI01000832 JAI16771.1 NNAY01000782 OXU26536.1 AJVK01008965 GFDF01008109 JAV05975.1 GEBQ01017855 JAT22122.1 KQ434796 KZC05534.1 ATLV01023219 KE525341 KFB48635.1 GFDG01002166 JAV16633.1 KQ435834 KOX71605.1 CH916367 EDW02491.1 CP012524 ALC40539.1 KA648054 AFP62683.1 GAMC01015718 JAB90837.1 GBXI01003575 JAD10717.1 CM000071 EAL25695.1 CH902619 EDV36431.1 CH940662 EDW58797.1 PYGN01000399 PSN47026.1 CH933808 EDW08160.1 OUUW01000001 SPP74430.1 CM000157 EDW89454.1 CH954177 EDV59420.1 BT126144 KF051019 KX531362 ADZ74171.1 AHX83805.1 ANY27172.1 AF173550 AE013599 AY071005 AAF58987.1 AAL29185.1 AAL48627.1 CH479183 EDW36112.1 GDHF01013886 JAI38428.1 GAKP01013182 JAC45770.1 CH964282 EDW85776.1 CH480816 EDW47048.1 CM000362 CM002911 EDX06294.1 KMY92433.1 JRES01000265 KNC32827.1 CCAG010004671 EZ421943 EZ424294 ADD18239.1 ADD20570.1 JXJN01016042 GGLE01001633 MBY05759.1 HM132050 AEC22816.1 UFQS01000305 UFQT01000305 SSX02680.1 SSX23054.1 IACF01000981 LAB66714.1 CVRI01000039 CRK94618.1
KQ460890 KPJ10979.1 NWSH01002905 PCG67330.1 ODYU01006665 SOQ48710.1 GU108331 ADK56161.1 GAIX01000063 JAA92497.1 AK401978 KQ459593 BAM18600.1 KPI96557.1 GDQN01008331 GDQN01004536 GDQN01001903 JAT82723.1 JAT86518.1 JAT89151.1 AGBW02010065 OWR49266.1 GDQN01008797 GDQN01002866 JAT82257.1 JAT88188.1 JXUM01011038 KQ560322 KXJ83062.1 AXCN02000449 AXCM01001406 CH477404 EAT41626.1 KQ979709 KYN19479.1 ADTU01006397 GGFM01005016 MBW25767.1 GGFK01009749 MBW43070.1 GGFJ01010061 MBW59202.1 ADMH02001414 ETN62618.1 KQ982138 KYQ59605.1 GFDL01011273 JAV23772.1 AAAB01008960 EAA11513.3 APCN01005863 GL434886 EFN74382.1 KK107024 QOIP01000004 EZA62220.1 RLU23391.1 KQ982450 KYQ56264.1 LBMM01006642 KMQ90437.1 GL888505 EGI59886.1 KQ976633 KYM78876.1 KQ981928 KYN32879.1 KQ977771 KYN00006.1 DS231969 EDS29698.1 GL451846 EFN78442.1 KC740978 AGM32802.1 NEVH01007821 PNF35367.1 GECU01012636 JAS95070.1 KQ414784 KOC61081.1 GANO01000471 JAB59400.1 GL447410 EFN86470.1 GECZ01012805 JAS56964.1 GEDC01008649 JAS28649.1 KK852897 KDR14174.1 GDAI01000832 JAI16771.1 NNAY01000782 OXU26536.1 AJVK01008965 GFDF01008109 JAV05975.1 GEBQ01017855 JAT22122.1 KQ434796 KZC05534.1 ATLV01023219 KE525341 KFB48635.1 GFDG01002166 JAV16633.1 KQ435834 KOX71605.1 CH916367 EDW02491.1 CP012524 ALC40539.1 KA648054 AFP62683.1 GAMC01015718 JAB90837.1 GBXI01003575 JAD10717.1 CM000071 EAL25695.1 CH902619 EDV36431.1 CH940662 EDW58797.1 PYGN01000399 PSN47026.1 CH933808 EDW08160.1 OUUW01000001 SPP74430.1 CM000157 EDW89454.1 CH954177 EDV59420.1 BT126144 KF051019 KX531362 ADZ74171.1 AHX83805.1 ANY27172.1 AF173550 AE013599 AY071005 AAF58987.1 AAL29185.1 AAL48627.1 CH479183 EDW36112.1 GDHF01013886 JAI38428.1 GAKP01013182 JAC45770.1 CH964282 EDW85776.1 CH480816 EDW47048.1 CM000362 CM002911 EDX06294.1 KMY92433.1 JRES01000265 KNC32827.1 CCAG010004671 EZ421943 EZ424294 ADD18239.1 ADD20570.1 JXJN01016042 GGLE01001633 MBY05759.1 HM132050 AEC22816.1 UFQS01000305 UFQT01000305 SSX02680.1 SSX23054.1 IACF01000981 LAB66714.1 CVRI01000039 CRK94618.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000007151
UP000069940
UP000249989
+ More
UP000075886 UP000075884 UP000075883 UP000075900 UP000075885 UP000008820 UP000076408 UP000078492 UP000005205 UP000000673 UP000069272 UP000075809 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000311 UP000053097 UP000279307 UP000075880 UP000036403 UP000007755 UP000078540 UP000078541 UP000078542 UP000002320 UP000002358 UP000008237 UP000235965 UP000053825 UP000027135 UP000215335 UP000092462 UP000076502 UP000030765 UP000053105 UP000001070 UP000092553 UP000095301 UP000095300 UP000001819 UP000007801 UP000008792 UP000245037 UP000009192 UP000268350 UP000002282 UP000008711 UP000000803 UP000192221 UP000008744 UP000007798 UP000075920 UP000001292 UP000000304 UP000037069 UP000091820 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000183832
UP000075886 UP000075884 UP000075883 UP000075900 UP000075885 UP000008820 UP000076408 UP000078492 UP000005205 UP000000673 UP000069272 UP000075809 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000311 UP000053097 UP000279307 UP000075880 UP000036403 UP000007755 UP000078540 UP000078541 UP000078542 UP000002320 UP000002358 UP000008237 UP000235965 UP000053825 UP000027135 UP000215335 UP000092462 UP000076502 UP000030765 UP000053105 UP000001070 UP000092553 UP000095301 UP000095300 UP000001819 UP000007801 UP000008792 UP000245037 UP000009192 UP000268350 UP000002282 UP000008711 UP000000803 UP000192221 UP000008744 UP000007798 UP000075920 UP000001292 UP000000304 UP000037069 UP000091820 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000183832
Interpro
IPR008111
RNA-bd_8
+ More
IPR033744 RRM_RBM8
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR008733 PEX11
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011020 HTTM
IPR011527 ABC1_TM_dom
IPR007782 VKG_COase
IPR017871 ABC_transporter_CS
IPR039421 Type_I_exporter
IPR003593 AAA+_ATPase
IPR003439 ABC_transporter-like
IPR033744 RRM_RBM8
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR008733 PEX11
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011020 HTTM
IPR011527 ABC1_TM_dom
IPR007782 VKG_COase
IPR017871 ABC_transporter_CS
IPR039421 Type_I_exporter
IPR003593 AAA+_ATPase
IPR003439 ABC_transporter-like
Gene 3D
CDD
ProteinModelPortal
Q1HQB4
A0A194R0C8
A0A2A4J7A8
A0A2H1W6J9
G8A442
S4Q0B8
+ More
I4DL10 A0A1E1W6X4 A0A212F6A2 A0A1E1W5J3 A0A182GH26 A0A182QXI2 A0A182NJA3 A0A182MMH8 A0A1I8JUP7 A0A182PEQ9 Q175A5 A0A182YR35 A0A195E3N0 A0A158P1E8 A0A2M3ZB47 A0A2M4AQL4 A0A2M4C1L7 W5JH19 A0A182FCB2 A0A151XH13 A0A1Q3F892 A0A182UND7 A0A182K4Q3 A0A182UL70 A0A182WTS0 A0A182LL96 Q7Q5L6 A0A182HI17 E1ZWQ4 A0A026X1P1 A0A182J1W8 A0A151X7G2 A0A0J7KJJ2 F4X0Y4 A0A151I000 A0A195EYB5 A0A151IFT1 B0WKA7 K7J3K6 E2C0P8 R4V253 A0A2J7R3F3 A0A1B6J7C5 A0A0L7QRF4 U5EYB1 E2BCV8 A0A1B6G3G0 A0A1B6DSP1 A0A067QXR0 A0A0K8TRK4 A0A232F6P0 A0A1B0DR84 A0A1L8DHX2 A0A1B6LEK0 A0A154P2G6 A0A084WEJ1 A0A1L8EDI0 A0A0M8ZUV4 B4J9I4 A0A0M3QUF4 T1PH98 A0A1I8PAK5 W8AXF2 A0A0A1XID3 Q28ZB8 B3MDP6 B4ME32 A0A2P8YS07 B4KQJ1 A0A3B0JGF2 B4P3X1 B3N7L5 F2FB57 Q9V535 A0A1W4UZM2 B4GHZ2 A0A0K8VHU1 A0A034VVB5 B4NN37 A0A182WCC1 B4HSM1 B4QGR9 A0A0L0CKR0 A0A1A9W0Y8 A0A1A9UU10 A0A1A9ZUX9 D3TKL1 A0A1A9XY30 A0A1B0BKU7 A0A2R5L8F6 M9MSS7 A0A336KNV2 A0A2P2HY58 A0A1J1I4D2
I4DL10 A0A1E1W6X4 A0A212F6A2 A0A1E1W5J3 A0A182GH26 A0A182QXI2 A0A182NJA3 A0A182MMH8 A0A1I8JUP7 A0A182PEQ9 Q175A5 A0A182YR35 A0A195E3N0 A0A158P1E8 A0A2M3ZB47 A0A2M4AQL4 A0A2M4C1L7 W5JH19 A0A182FCB2 A0A151XH13 A0A1Q3F892 A0A182UND7 A0A182K4Q3 A0A182UL70 A0A182WTS0 A0A182LL96 Q7Q5L6 A0A182HI17 E1ZWQ4 A0A026X1P1 A0A182J1W8 A0A151X7G2 A0A0J7KJJ2 F4X0Y4 A0A151I000 A0A195EYB5 A0A151IFT1 B0WKA7 K7J3K6 E2C0P8 R4V253 A0A2J7R3F3 A0A1B6J7C5 A0A0L7QRF4 U5EYB1 E2BCV8 A0A1B6G3G0 A0A1B6DSP1 A0A067QXR0 A0A0K8TRK4 A0A232F6P0 A0A1B0DR84 A0A1L8DHX2 A0A1B6LEK0 A0A154P2G6 A0A084WEJ1 A0A1L8EDI0 A0A0M8ZUV4 B4J9I4 A0A0M3QUF4 T1PH98 A0A1I8PAK5 W8AXF2 A0A0A1XID3 Q28ZB8 B3MDP6 B4ME32 A0A2P8YS07 B4KQJ1 A0A3B0JGF2 B4P3X1 B3N7L5 F2FB57 Q9V535 A0A1W4UZM2 B4GHZ2 A0A0K8VHU1 A0A034VVB5 B4NN37 A0A182WCC1 B4HSM1 B4QGR9 A0A0L0CKR0 A0A1A9W0Y8 A0A1A9UU10 A0A1A9ZUX9 D3TKL1 A0A1A9XY30 A0A1B0BKU7 A0A2R5L8F6 M9MSS7 A0A336KNV2 A0A2P2HY58 A0A1J1I4D2
PDB
2X1G
E-value=2.36741e-52,
Score=514
Ontologies
PATHWAY
GO
GO:0008380
GO:0005737
GO:0051028
GO:0003729
GO:0016607
GO:0006397
GO:0000184
GO:0035145
GO:0000381
GO:0000226
GO:0007317
GO:0051663
GO:0007294
GO:0000335
GO:0007314
GO:0005779
GO:0016559
GO:0071013
GO:0005634
GO:0045451
GO:0046595
GO:0000398
GO:0007310
GO:0008488
GO:0005524
GO:0042626
GO:0017187
GO:0016021
GO:0003676
GO:0003723
GO:0006396
GO:0000166
GO:0004887
GO:0003707
PANTHER
Topology
Subcellular location
Nucleus
Nucleus speckle
Cytoplasm
Nucleus speckle
Cytoplasm
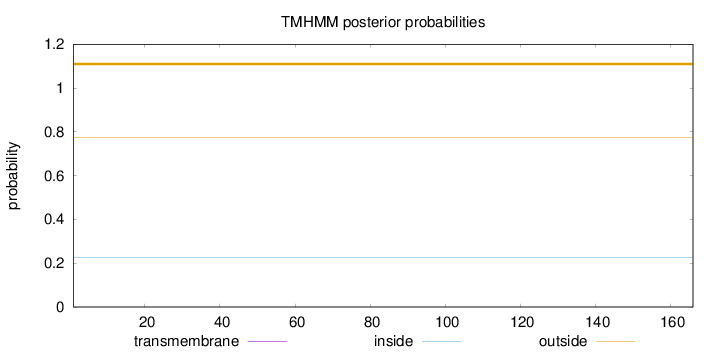
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00135
Exp number, first 60 AAs:
0
Total prob of N-in:
0.22679
outside
1 - 166
Population Genetic Test Statistics
Pi
291.002789
Theta
170.11937
Tajima's D
2.289611
CLR
0.213658
CSRT
0.91870406479676
Interpretation
Uncertain
