Pre Gene Modal
BGIBMGA009684
Annotation
PREDICTED:_proteasome_subunit_beta_type-4_[Amyelois_transitella]
Full name
Proteasome subunit beta
Location in the cell
Cytoplasmic Reliability : 1.176
Sequence
CDS
ATGGCTTTTATGGGGGACACGATGAACCCCACGCCGTTGTGGCAAAACGGACCGTCGCCTGGAGCATTTTATAATTTTCCTGGAAATGCTTCCACAATTGCTCCATCCAGGCACGGTGTACAGGATTTTACAGCTCACTCAGCGAGCCCCATCACGACCACCACAACTGTCATCGGAGTTAAGTTTGACAAGGGATGTGTTATCGCCGGTGACACACTGGGCTCGTACGGGTCATTGGCCCGGTTTCGAGACTGTCCCCGCGTGATGAAAGTCAATGACTTAATACTACTAGGCTGCGGAGGAGACTATGCTGATTTTCAGTATTTGAAGGATATCATTCAACAAAAAATCATTGATGAGCGCTGTGTTGGAGATGGTCTCCAGCTCAAGCCCCGTTCTCTGCACTGCTGGTTGACTCGTGTCCTCTACAACAAGAGGAGCAAGATGGATCCCCTCTGGAACAGCTATGTGGTTGCCGGTATTCAGGATGGTGAACCCTTTCTCGGAGCTGTTGATAAATTGGGAACAGCCTATGAAGATGCTGTGATCTCGAACGGGCTAGGAGCTTACATGGCGACTCCTCTACTGAGGGACGCAGTGGATAAAGGACCTCTGGACCAAGAAACTGCTATTGCGGTTGTACGTAAAAGTATGGAGGTGCTTTTCTACAGAGATGCTCGCGCGTTCCAACGCTACCAGCTCGGCGTGGTCACGGCTGAGGGAATAAAGATCACAGACGAGGAGCTCAAGCATGATTGGTCGTTGGCTCACATGATACAAATGAAGTGA
Protein
MAFMGDTMNPTPLWQNGPSPGAFYNFPGNASTIAPSRHGVQDFTAHSASPITTTTTVIGVKFDKGCVIAGDTLGSYGSLARFRDCPRVMKVNDLILLGCGGDYADFQYLKDIIQQKIIDERCVGDGLQLKPRSLHCWLTRVLYNKRSKMDPLWNSYVVAGIQDGEPFLGAVDKLGTAYEDAVISNGLGAYMATPLLRDAVDKGPLDQETAIAVVRKSMEVLFYRDARAFQRYQLGVVTAEGIKITDEELKHDWSLAHMIQMK
Summary
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
Similarity
Belongs to the peptidase T1B family.
Uniprot
H9JJI3
A0A2H1W6J7
A0A2A4J617
A0A1E1WDG0
S4P1S9
A0A194QZU6
+ More
I4DL87 A0A194PZG5 A0A1Y1MEW2 D6WTK4 A0A2P8YT67 A0A1B6GG72 A0A1B6K4B3 K7IPQ8 A0A232ERM7 A0A067QRU4 A0A1B6MSW4 E2AWR1 A0A0J7KKX3 F4WYJ3 A0A0L7RCZ9 E2BNX4 A0A195E3I7 A0A195F4G4 A0A1W4WKB4 A0A154P3I6 A0A151I2H5 A0A151K235 A0A310SJ20 A0A0M9AA60 A0A088AM79 A0A2A3EHE4 V9IL75 T1JMI3 A0A158P1G3 A0A151X7C1 V4A971 A0A0B7AYI7 A0A0L7LH29 E9G7M2 A0A210PN58 A0A3L8DVQ0 C3YDQ6 A0A1W5APG6 A0A0T6B449 A0A0P5Z2Y6 Q4SZT8 H3CIJ2 A0A1A8CY81 A0A1A8AVF5 A0A0P5FC83 A0A060XUI2 C1BGS6 A0A3B3XY46 A0A1S3PYJ0 A0A1B6CU75 D6R733 B5XFR6 A0A0P5HWG2 A0A3P9PRR1 A0A3P8YM14 A0A087XXP5 A0A3B3VXI3 A0A2T7PS28 A0A1S3JE44 A0A1A8SDP9 B5X5F6 A0A1S3JDE7 W5LMS4 H2RJK8 A0A2J7PD67 A8E7N8 A0A1A8IAE9 B5X7W0 A0A2I4CKW4 A0A3P8RYR9 N6TI04 A0A1A8M6R9 H3A4V7 B7NZD7 A0A0K8RM43 H3A4V6 A0A060VQY7 A0A1A8FV47 B5X9D0 A0A3P9CU85 Q90ZQ5 G1T918 A0A3B3QWW3 A0A2D0PWD6 E3TBP4 A0A131YAH7 A0A2U3W7W6 A0A146NLI0 A0A146MHN3 D2HBR7 J3JY13 G1L1C8 E0VBP9
I4DL87 A0A194PZG5 A0A1Y1MEW2 D6WTK4 A0A2P8YT67 A0A1B6GG72 A0A1B6K4B3 K7IPQ8 A0A232ERM7 A0A067QRU4 A0A1B6MSW4 E2AWR1 A0A0J7KKX3 F4WYJ3 A0A0L7RCZ9 E2BNX4 A0A195E3I7 A0A195F4G4 A0A1W4WKB4 A0A154P3I6 A0A151I2H5 A0A151K235 A0A310SJ20 A0A0M9AA60 A0A088AM79 A0A2A3EHE4 V9IL75 T1JMI3 A0A158P1G3 A0A151X7C1 V4A971 A0A0B7AYI7 A0A0L7LH29 E9G7M2 A0A210PN58 A0A3L8DVQ0 C3YDQ6 A0A1W5APG6 A0A0T6B449 A0A0P5Z2Y6 Q4SZT8 H3CIJ2 A0A1A8CY81 A0A1A8AVF5 A0A0P5FC83 A0A060XUI2 C1BGS6 A0A3B3XY46 A0A1S3PYJ0 A0A1B6CU75 D6R733 B5XFR6 A0A0P5HWG2 A0A3P9PRR1 A0A3P8YM14 A0A087XXP5 A0A3B3VXI3 A0A2T7PS28 A0A1S3JE44 A0A1A8SDP9 B5X5F6 A0A1S3JDE7 W5LMS4 H2RJK8 A0A2J7PD67 A8E7N8 A0A1A8IAE9 B5X7W0 A0A2I4CKW4 A0A3P8RYR9 N6TI04 A0A1A8M6R9 H3A4V7 B7NZD7 A0A0K8RM43 H3A4V6 A0A060VQY7 A0A1A8FV47 B5X9D0 A0A3P9CU85 Q90ZQ5 G1T918 A0A3B3QWW3 A0A2D0PWD6 E3TBP4 A0A131YAH7 A0A2U3W7W6 A0A146NLI0 A0A146MHN3 D2HBR7 J3JY13 G1L1C8 E0VBP9
EC Number
3.4.25.1
Pubmed
19121390
23622113
26354079
22651552
28004739
18362917
+ More
19820115 29403074 20075255 28648823 24845553 20798317 21719571 21347285 23254933 26227816 21292972 28812685 30249741 18563158 15496914 24755649 20433749 25069045 25329095 21551351 23594743 23537049 9215903 25186727 12457246 21993624 29240929 20634964 20010809 22516182 20566863
19820115 29403074 20075255 28648823 24845553 20798317 21719571 21347285 23254933 26227816 21292972 28812685 30249741 18563158 15496914 24755649 20433749 25069045 25329095 21551351 23594743 23537049 9215903 25186727 12457246 21993624 29240929 20634964 20010809 22516182 20566863
EMBL
BABH01017541
ODYU01006665
SOQ48709.1
NWSH01002905
PCG67329.1
GDQN01006153
+ More
JAT84901.1 GAIX01012335 JAA80225.1 KQ460890 KPJ10977.1 AK402055 BAM18677.1 KQ459593 KPI96555.1 GEZM01037901 JAV81857.1 KQ971354 EFA05833.1 PYGN01000374 PSN47440.1 GECZ01008363 JAS61406.1 GECU01001420 JAT06287.1 NNAY01002575 OXU21015.1 KK853011 KDR12542.1 GEBQ01000945 JAT39032.1 GL443403 EFN62137.1 LBMM01006160 KMQ90871.1 GL888450 EGI60725.1 KQ414615 KOC68670.1 GL449511 EFN82566.1 KQ979709 KYN19444.1 KQ981852 KYN34974.1 KQ434796 KZC05798.1 KQ976538 KYM81319.1 LKEX01009667 KYN50132.1 KQ762882 OAD55382.1 KQ435711 KOX79626.1 KZ288248 PBC31130.1 JR053230 AEY61883.1 JH431845 ADTU01006535 KQ982450 KYQ56271.1 KB200701 ESP00519.1 HACG01038983 CEK85848.1 JTDY01001158 KOB74710.1 GL732534 EFX84494.1 NEDP02005576 OWF37903.1 QOIP01000004 RLU24019.1 GG666504 EEN61525.1 LJIG01016021 KRT81853.1 GDIP01050016 LRGB01002076 JAM53699.1 KZS09320.1 CAAE01011492 CAF93844.1 HADZ01019854 HAEA01001890 SBP83795.1 HADY01020183 HAEJ01004842 SBP58668.1 GDIQ01257574 JAJ94150.1 FR905650 CDQ80754.1 BT073805 ACO08229.1 GEDC01020341 JAS16957.1 HM124771 ADF97632.1 BT049885 ACI69686.1 GDIQ01224114 JAK27611.1 AYCK01011407 PZQS01000002 PVD36233.1 HAEH01014303 HAEI01013310 SBS15779.1 BT046275 ACI66076.1 NEVH01026393 PNF14268.1 BX571688 HAED01007870 HAEE01013504 SBQ94082.1 BT047129 ACI66930.1 APGK01025883 KB740605 ENN80034.1 HAEF01011539 HAEG01010879 SBR52532.1 AFYH01247314 DP001034 ACJ74020.1 GADI01001653 JAA72155.1 FR904286 CDQ57373.1 HAEB01015467 HAEC01001930 SBQ61994.1 BT047649 ACI67450.1 AF361366 AAK51461.1 AAGW02000574 GU587768 ADO27730.1 GEFM01000346 JAP75450.1 GCES01154047 JAQ32275.1 GCES01167119 JAQ19203.1 GL192667 EFB28453.1 BT128136 AEE63097.1 ACTA01000202 DS235035 EEB10805.1
JAT84901.1 GAIX01012335 JAA80225.1 KQ460890 KPJ10977.1 AK402055 BAM18677.1 KQ459593 KPI96555.1 GEZM01037901 JAV81857.1 KQ971354 EFA05833.1 PYGN01000374 PSN47440.1 GECZ01008363 JAS61406.1 GECU01001420 JAT06287.1 NNAY01002575 OXU21015.1 KK853011 KDR12542.1 GEBQ01000945 JAT39032.1 GL443403 EFN62137.1 LBMM01006160 KMQ90871.1 GL888450 EGI60725.1 KQ414615 KOC68670.1 GL449511 EFN82566.1 KQ979709 KYN19444.1 KQ981852 KYN34974.1 KQ434796 KZC05798.1 KQ976538 KYM81319.1 LKEX01009667 KYN50132.1 KQ762882 OAD55382.1 KQ435711 KOX79626.1 KZ288248 PBC31130.1 JR053230 AEY61883.1 JH431845 ADTU01006535 KQ982450 KYQ56271.1 KB200701 ESP00519.1 HACG01038983 CEK85848.1 JTDY01001158 KOB74710.1 GL732534 EFX84494.1 NEDP02005576 OWF37903.1 QOIP01000004 RLU24019.1 GG666504 EEN61525.1 LJIG01016021 KRT81853.1 GDIP01050016 LRGB01002076 JAM53699.1 KZS09320.1 CAAE01011492 CAF93844.1 HADZ01019854 HAEA01001890 SBP83795.1 HADY01020183 HAEJ01004842 SBP58668.1 GDIQ01257574 JAJ94150.1 FR905650 CDQ80754.1 BT073805 ACO08229.1 GEDC01020341 JAS16957.1 HM124771 ADF97632.1 BT049885 ACI69686.1 GDIQ01224114 JAK27611.1 AYCK01011407 PZQS01000002 PVD36233.1 HAEH01014303 HAEI01013310 SBS15779.1 BT046275 ACI66076.1 NEVH01026393 PNF14268.1 BX571688 HAED01007870 HAEE01013504 SBQ94082.1 BT047129 ACI66930.1 APGK01025883 KB740605 ENN80034.1 HAEF01011539 HAEG01010879 SBR52532.1 AFYH01247314 DP001034 ACJ74020.1 GADI01001653 JAA72155.1 FR904286 CDQ57373.1 HAEB01015467 HAEC01001930 SBQ61994.1 BT047649 ACI67450.1 AF361366 AAK51461.1 AAGW02000574 GU587768 ADO27730.1 GEFM01000346 JAP75450.1 GCES01154047 JAQ32275.1 GCES01167119 JAQ19203.1 GL192667 EFB28453.1 BT128136 AEE63097.1 ACTA01000202 DS235035 EEB10805.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007266
UP000245037
+ More
UP000002358 UP000215335 UP000027135 UP000000311 UP000036403 UP000007755 UP000053825 UP000008237 UP000078492 UP000078541 UP000192223 UP000076502 UP000078540 UP000078542 UP000053105 UP000005203 UP000242457 UP000005205 UP000075809 UP000030746 UP000037510 UP000000305 UP000242188 UP000279307 UP000001554 UP000192224 UP000076858 UP000007303 UP000193380 UP000261480 UP000087266 UP000242638 UP000265140 UP000028760 UP000261500 UP000245119 UP000085678 UP000018467 UP000005226 UP000235965 UP000000437 UP000192220 UP000265080 UP000019118 UP000008672 UP000265160 UP000001811 UP000261540 UP000221080 UP000245340 UP000008912 UP000009046
UP000002358 UP000215335 UP000027135 UP000000311 UP000036403 UP000007755 UP000053825 UP000008237 UP000078492 UP000078541 UP000192223 UP000076502 UP000078540 UP000078542 UP000053105 UP000005203 UP000242457 UP000005205 UP000075809 UP000030746 UP000037510 UP000000305 UP000242188 UP000279307 UP000001554 UP000192224 UP000076858 UP000007303 UP000193380 UP000261480 UP000087266 UP000242638 UP000265140 UP000028760 UP000261500 UP000245119 UP000085678 UP000018467 UP000005226 UP000235965 UP000000437 UP000192220 UP000265080 UP000019118 UP000008672 UP000265160 UP000001811 UP000261540 UP000221080 UP000245340 UP000008912 UP000009046
PRIDE
Pfam
PF00227 Proteasome
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
H9JJI3
A0A2H1W6J7
A0A2A4J617
A0A1E1WDG0
S4P1S9
A0A194QZU6
+ More
I4DL87 A0A194PZG5 A0A1Y1MEW2 D6WTK4 A0A2P8YT67 A0A1B6GG72 A0A1B6K4B3 K7IPQ8 A0A232ERM7 A0A067QRU4 A0A1B6MSW4 E2AWR1 A0A0J7KKX3 F4WYJ3 A0A0L7RCZ9 E2BNX4 A0A195E3I7 A0A195F4G4 A0A1W4WKB4 A0A154P3I6 A0A151I2H5 A0A151K235 A0A310SJ20 A0A0M9AA60 A0A088AM79 A0A2A3EHE4 V9IL75 T1JMI3 A0A158P1G3 A0A151X7C1 V4A971 A0A0B7AYI7 A0A0L7LH29 E9G7M2 A0A210PN58 A0A3L8DVQ0 C3YDQ6 A0A1W5APG6 A0A0T6B449 A0A0P5Z2Y6 Q4SZT8 H3CIJ2 A0A1A8CY81 A0A1A8AVF5 A0A0P5FC83 A0A060XUI2 C1BGS6 A0A3B3XY46 A0A1S3PYJ0 A0A1B6CU75 D6R733 B5XFR6 A0A0P5HWG2 A0A3P9PRR1 A0A3P8YM14 A0A087XXP5 A0A3B3VXI3 A0A2T7PS28 A0A1S3JE44 A0A1A8SDP9 B5X5F6 A0A1S3JDE7 W5LMS4 H2RJK8 A0A2J7PD67 A8E7N8 A0A1A8IAE9 B5X7W0 A0A2I4CKW4 A0A3P8RYR9 N6TI04 A0A1A8M6R9 H3A4V7 B7NZD7 A0A0K8RM43 H3A4V6 A0A060VQY7 A0A1A8FV47 B5X9D0 A0A3P9CU85 Q90ZQ5 G1T918 A0A3B3QWW3 A0A2D0PWD6 E3TBP4 A0A131YAH7 A0A2U3W7W6 A0A146NLI0 A0A146MHN3 D2HBR7 J3JY13 G1L1C8 E0VBP9
I4DL87 A0A194PZG5 A0A1Y1MEW2 D6WTK4 A0A2P8YT67 A0A1B6GG72 A0A1B6K4B3 K7IPQ8 A0A232ERM7 A0A067QRU4 A0A1B6MSW4 E2AWR1 A0A0J7KKX3 F4WYJ3 A0A0L7RCZ9 E2BNX4 A0A195E3I7 A0A195F4G4 A0A1W4WKB4 A0A154P3I6 A0A151I2H5 A0A151K235 A0A310SJ20 A0A0M9AA60 A0A088AM79 A0A2A3EHE4 V9IL75 T1JMI3 A0A158P1G3 A0A151X7C1 V4A971 A0A0B7AYI7 A0A0L7LH29 E9G7M2 A0A210PN58 A0A3L8DVQ0 C3YDQ6 A0A1W5APG6 A0A0T6B449 A0A0P5Z2Y6 Q4SZT8 H3CIJ2 A0A1A8CY81 A0A1A8AVF5 A0A0P5FC83 A0A060XUI2 C1BGS6 A0A3B3XY46 A0A1S3PYJ0 A0A1B6CU75 D6R733 B5XFR6 A0A0P5HWG2 A0A3P9PRR1 A0A3P8YM14 A0A087XXP5 A0A3B3VXI3 A0A2T7PS28 A0A1S3JE44 A0A1A8SDP9 B5X5F6 A0A1S3JDE7 W5LMS4 H2RJK8 A0A2J7PD67 A8E7N8 A0A1A8IAE9 B5X7W0 A0A2I4CKW4 A0A3P8RYR9 N6TI04 A0A1A8M6R9 H3A4V7 B7NZD7 A0A0K8RM43 H3A4V6 A0A060VQY7 A0A1A8FV47 B5X9D0 A0A3P9CU85 Q90ZQ5 G1T918 A0A3B3QWW3 A0A2D0PWD6 E3TBP4 A0A131YAH7 A0A2U3W7W6 A0A146NLI0 A0A146MHN3 D2HBR7 J3JY13 G1L1C8 E0VBP9
PDB
6MSK
E-value=1.6352e-69,
Score=665
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
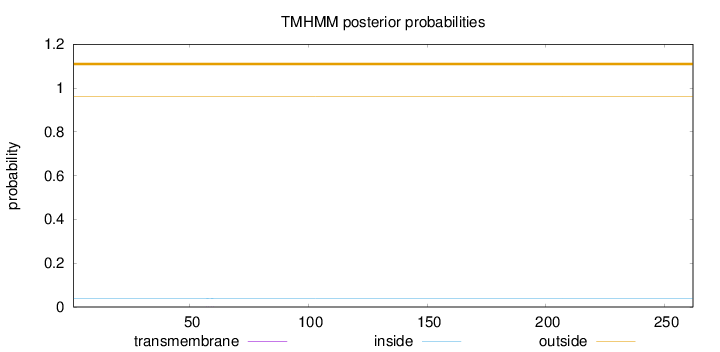
Length:
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00778999999999999
Exp number, first 60 AAs:
0.00296
Total prob of N-in:
0.03853
outside
1 - 262
Population Genetic Test Statistics
Pi
195.829596
Theta
178.442056
Tajima's D
0.714383
CLR
12.131517
CSRT
0.584520773961302
Interpretation
Uncertain
