Gene
KWMTBOMO00834 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009685
Annotation
hypothetical_protein_KGM_20484_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.416
Sequence
CDS
ATGCAGAAACAAGTGCCTGCTAAAGGTAAAAAACGTAAAGCAAATACAAACTTAGTAACATCGGAACCTAAAAATGTGAAACAAAACTCTTTGGTTAACTCTGATAATAAAAATAGCATCTTAGATCCTAAAAATATTAACAAAAACGTTAATAAGAATAAAAATAAACAAAAAAATCAAAATACTGTTAATTTGGCACCTATTTTAGCCCAGTCTAATCTGGCAACTTCCACAGACAAAAATAACACTAAAAATGACACAAACATAAAAAAATTTGACAATTCTGGGAAGAACAAACAACAACAGAAATCAAAACCAACTCCGCTAGCAGAAAAAAACGTTATTGATGCAGGTGGAATTAAAATTGAAAACGACATTAAAAAAGAGACTGATTCGAATGCGGCTAAAAAAAAAAGGAGAAACAAGCGTAAAAAGATACCCGCACAAGCAACCTTGCTTAATGAAAATGGTATGAATGACTCGAGTGATGATGAAGACGTACCTGACAAAAGTATACCTCAAAAACAGACTCAGACTCAGAAACCTAAGAAAAATGTACTTAAAATAAATAATACACTGCCTGTCGTGCATAAAACTGAAAACGAAGCAGATGTGTCTAGTGACGATGAAGCCCCACCTGCAAATGACAAGAAGAGTGCCAACTTACCTGACGCAGTGCAGAAGAAGAAGAAAAATAAACGCAAGAAGAAGAATGCGGCGACCAAGGTTGAAATTTCAAATGGTAATGAATCTTCAAATAACCAAGGAGAAAAAACACCTCAACAAGCACTTAAAAGTAAGAAAAATAAACAAAACATTGCAAATGGCAAAAATACTGAAGAATCCAGTGAAGATGAAGACTCCAGTGAAAAACCTCAAACGATTATAAATCAAAACGGTAAAGCACAGTCATCAAATGCATCTTCAGCGTCTAGTGATGAAGAACCACCGAAAAAGGATGGTAAAGTCAAACTGAACAATAAAGATATAAAAACGGCCGCCTTGTTGAAGGTTAAGCATGAGGCAGACAAAATAGATATGTCCGAAAGATATCCCATATTGAAAGATCAAAAGTTTGTGGAATCTTTCATGAATGTTAAAATCACTAACGGTCAGAAAGGTCGCATAAGGCAAGCCTTGTTTAAACAGTTCAAAGACATACCAGACGAACAAAAACCAGACCTAATCCATGCGAGAATTCAAGCCATGATCAAAGGAACTATCATGAATGATTCACAATTACGCAGAATTCGGATTCTATATAATATGCTGAAAGAGACACTTAAGAAGCTGGGACACAAAACTCCTGTCGTAGAAAATTTAAGTAAAGAGAAAAAGCAAAAGGTTTCCAAAGAGCAAGAGAAGGTTAAGGGGCCAAAGAGATATGTGGTGTTTGTAGGAAATCTACCAGTGGGAATTCAAAAGGAGAGACTTACACATCATTTCTTAGACATAAGTGACAACATTAAGGACATCAGATTGCCTGAAACTCCAGAAGGCAAGAAAGCTGCAATTGCCTATATCGAACTAGATAATGAAACAAGTTATGAGCTGGCACTGTCAAAGAATCACACAATGTTGGACAACAGGAGAATCAACGTACAGTATTCCGTGCAGAAGAACAGTAAAATAACATTAACCGAAGCCAAGGGTAAATCAGCGAAACTGTTGGCCTTACAGAAAGCTGGAAAGCTTATCGGTAGCGTCCCGCTGGAGCAGAAGCGAAGTCAACGTCGTAACAAGATGAAAAAAATGCGCGCTGAAGCCGAAGCTACGGCGTGA
Protein
MQKQVPAKGKKRKANTNLVTSEPKNVKQNSLVNSDNKNSILDPKNINKNVNKNKNKQKNQNTVNLAPILAQSNLATSTDKNNTKNDTNIKKFDNSGKNKQQQKSKPTPLAEKNVIDAGGIKIENDIKKETDSNAAKKKRRNKRKKIPAQATLLNENGMNDSSDDEDVPDKSIPQKQTQTQKPKKNVLKINNTLPVVHKTENEADVSSDDEAPPANDKKSANLPDAVQKKKKNKRKKKNAATKVEISNGNESSNNQGEKTPQQALKSKKNKQNIANGKNTEESSEDEDSSEKPQTIINQNGKAQSSNASSASSDEEPPKKDGKVKLNNKDIKTAALLKVKHEADKIDMSERYPILKDQKFVESFMNVKITNGQKGRIRQALFKQFKDIPDEQKPDLIHARIQAMIKGTIMNDSQLRRIRILYNMLKETLKKLGHKTPVVENLSKEKKQKVSKEQEKVKGPKRYVVFVGNLPVGIQKERLTHHFLDISDNIKDIRLPETPEGKKAAIAYIELDNETSYELALSKNHTMLDNRRINVQYSVQKNSKITLTEAKGKSAKLLALQKAGKLIGSVPLEQKRSQRRNKMKKMRAEAEATA
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Pfam
PF00076 RRM_1
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
PDB
2DB1
E-value=0.0321402,
Score=89
Ontologies
GO
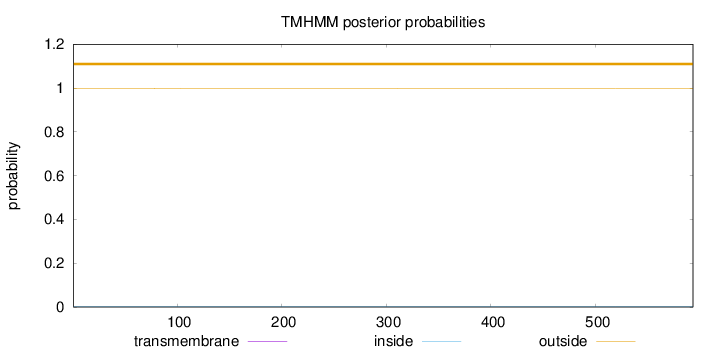
Topology
Length:
593
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00122
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.00246
outside
1 - 593
Population Genetic Test Statistics
Pi
311.842664
Theta
181.588509
Tajima's D
2.133647
CLR
0.011212
CSRT
0.899705014749263
Interpretation
Uncertain
