Gene
KWMTBOMO00826
Pre Gene Modal
BGIBMGA009690
Annotation
PREDICTED:_androgen-dependent_TFPI-regulating_protein-like_[Bombyx_mori]
Full name
Androgen-dependent TFPI-regulating protein
Location in the cell
PlasmaMembrane Reliability : 4.69
Sequence
CDS
ATGAGCTATCAAATTTACTTTCGCTTGCTCGGCTATGTGTCGACAATAGCGATGCATATCGGTAATCTAATTTACATGCGGCATACCATACTATCGGCTAATATCAGCGAAAAATCCCTGGATAATGTCGGAATAGGCACTTTCAAGAAAATGGAGTATCGATATTTTACTACTTGGACGGTGGGTCTACAGATAGGATATGCGGCGCTGGGTACACTCTGCGATCTTCTGGTGCTACTAAATAAACACAAAAAGAACTACAGATTACCTAAATATTTGGGCTCGTCGCGGGAAATTATGTTCGCTGGTCTGGTATGGCCGTCTACTTGGGTAGTCTTCACTGTATTTTGGACTTTATTCAACTACGACAGAAGCCTGATATTCCCGGCTTACTTGGACAAGATCGTGACGCGTCTGTCTAACCACGTCATACATACAATGATCGTTCCGGTCGTGGTCTGGGAGGTGATGTTTCGGCCAAGGAAACCAAAAAGTCACTACAGGAACATCGCGCAACTGGCGATACACATGTTGGCGTACGTCTTCGTGTTACATTTCACTTATCACGAGCACGGCGTCTGGCTGTACCCGATCTTCAAAGCAGTTCAAGGAACGATACATTTCTATCTGCTCCACTTTTATGTCGCAGTTATGTGCTTGATGTTCTACTGGCTTCAATGGCGGTTGAACTCTTTGATCCACGGCGTCGAAGACGTAAAGAAGATTCACTGA
Protein
MSYQIYFRLLGYVSTIAMHIGNLIYMRHTILSANISEKSLDNVGIGTFKKMEYRYFTTWTVGLQIGYAALGTLCDLLVLLNKHKKNYRLPKYLGSSREIMFAGLVWPSTWVVFTVFWTLFNYDRSLIFPAYLDKIVTRLSNHVIHTMIVPVVVWEVMFRPRKPKSHYRNIAQLAIHMLAYVFVLHFTYHEHGVWLYPIFKAVQGTIHFYLLHFYVAVMCLMFYWLQWRLNSLIHGVEDVKKIH
Summary
Description
Regulates the expression and the cell-associated anticoagulant activity of the inhibitor TFPI in endothelial cells (in vitro).
Similarity
Belongs to the AIG1 family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Membrane
Polymorphism
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Androgen-dependent TFPI-regulating protein
splice variant In isoform 2.
sequence variant In dbSNP:rs2076185.
splice variant In isoform 2.
sequence variant In dbSNP:rs2076185.
Uniprot
H9JJI9
H9JJI8
S4P4C5
A0A2A4ISD3
A0A194R0B8
A0A194PSZ5
+ More
A0A212F7H0 A0A2A4ITZ9 A0A2H1WGM3 A0A194PTA5 I4DJC6 A0A194R142 A0A2A4J6I9 A0A1E1W9D6 A0A2H1WWJ5 A0A2A4JMB5 A0A2A4J503 A0A2A4ITN6 A0A2D4I7W0 A0A2D4GB73 T1IBD8 H9GE02 A0A1B6DTE3 A0A2K6D6X9 A0A2K6D6W2 A0A2J8WN06 A0A2I3N3A2 A0A2J7PDG0 A0A0D9R570 A0A2K5MJC8 A0A096NKY2 A0A2K5MJC9 H2PHY1 A0A2R9AV55 Q96IZ2 A0A2R9ASQ7 A0A2I3RF37 Q96IZ2-3 H9EXV2 A0A1D5Q198 A0A1B6CZW9 F7FVQ1 A0A341D0B8 G7P4F3 A0A2I3HSN3 G7MQK8 A0A2K6D6W5 G3S0A2 G3R864 A0A2K5JJ01 A0A2I3ML13 A0A2K6KYY2 A0A2K6RGG7 I3NHT6 A0A2K5MJD1 A0A2U4C5S4 M3XJ79 G3WW37 F7HLH0 A0A2K5WPU0 F6TKL5 A0A1S3MCH5 A0A060X0N9 F1RV85 M3WAX6 A0A2K5Y9N0 A0A1U7TW54 A0A2K5Y9N2 A0A2D0SL67 A0A2K5CIT5 A0A2K5WPV3 F7DN37 G1SDH9 A0A194PTG9 F7EH87 A0A2D0SL66 A0A151XB65 A0A067QU27 V8NRF1 A0A2K5JIW7 A0A2P8YZ85 A0A2K5RAU8 A0A2K6RGJ0 A0A2K6KZ03 F7I3Z6
A0A212F7H0 A0A2A4ITZ9 A0A2H1WGM3 A0A194PTA5 I4DJC6 A0A194R142 A0A2A4J6I9 A0A1E1W9D6 A0A2H1WWJ5 A0A2A4JMB5 A0A2A4J503 A0A2A4ITN6 A0A2D4I7W0 A0A2D4GB73 T1IBD8 H9GE02 A0A1B6DTE3 A0A2K6D6X9 A0A2K6D6W2 A0A2J8WN06 A0A2I3N3A2 A0A2J7PDG0 A0A0D9R570 A0A2K5MJC8 A0A096NKY2 A0A2K5MJC9 H2PHY1 A0A2R9AV55 Q96IZ2 A0A2R9ASQ7 A0A2I3RF37 Q96IZ2-3 H9EXV2 A0A1D5Q198 A0A1B6CZW9 F7FVQ1 A0A341D0B8 G7P4F3 A0A2I3HSN3 G7MQK8 A0A2K6D6W5 G3S0A2 G3R864 A0A2K5JJ01 A0A2I3ML13 A0A2K6KYY2 A0A2K6RGG7 I3NHT6 A0A2K5MJD1 A0A2U4C5S4 M3XJ79 G3WW37 F7HLH0 A0A2K5WPU0 F6TKL5 A0A1S3MCH5 A0A060X0N9 F1RV85 M3WAX6 A0A2K5Y9N0 A0A1U7TW54 A0A2K5Y9N2 A0A2D0SL67 A0A2K5CIT5 A0A2K5WPV3 F7DN37 G1SDH9 A0A194PTG9 F7EH87 A0A2D0SL66 A0A151XB65 A0A067QU27 V8NRF1 A0A2K5JIW7 A0A2P8YZ85 A0A2K5RAU8 A0A2K6RGJ0 A0A2K6KZ03 F7I3Z6
Pubmed
EMBL
BABH01017486
BABH01017487
BABH01017488
GAIX01006034
JAA86526.1
NWSH01008210
+ More
PCG62651.1 KQ460890 KPJ10969.1 KQ459593 KPI96546.1 AGBW02009872 OWR49681.1 PCG62652.1 ODYU01008534 SOQ52219.1 KPI96547.1 AK401394 BAM18016.1 KPJ10970.1 NWSH01002754 PCG67571.1 GDQN01007452 JAT83602.1 ODYU01011594 SOQ57431.1 NWSH01001079 PCG72724.1 NWSH01003298 PCG66594.1 NWSH01006591 PCG63347.1 IACK01084098 LAA80299.1 IACJ01112898 LAA57005.1 ACPB03005685 AAWZ02016749 AAWZ02016750 AAWZ02016751 GEDC01008352 JAS28946.1 NDHI03003384 PNJ71145.1 AHZZ02024143 NEVH01026389 PNF14352.1 AQIB01159356 ABGA01392235 ABGA01392236 ABGA01392237 ABGA01392238 ABGA01392239 ABGA01392240 ABGA01392241 ABGA01392242 AJFE02097058 AK313114 AK300919 AL022724 CH471087 BC007011 AACZ04065924 NBAG03000244 PNI62836.1 JU323455 AFE67211.1 JSUE03030254 JSUE03030255 GEDC01024428 GEDC01024128 GEDC01022375 GEDC01018863 GEDC01018435 GEDC01006808 JAS12870.1 JAS13170.1 JAS14923.1 JAS18435.1 JAS18863.1 JAS30490.1 CM001279 EHH52716.1 ADFV01015550 ADFV01015551 ADFV01015552 CM001256 EHH17956.1 CABD030043317 CABD030043318 CABD030043319 AGTP01048485 AGTP01048486 AGTP01048487 AFYH01049129 AFYH01049130 AFYH01049131 AFYH01049132 AEFK01063730 AEFK01063731 AEFK01063732 AEFK01063733 AEFK01063734 AEFK01063735 AQIA01052761 AQIA01052762 FR904873 CDQ73036.1 AEMK02000051 AANG04001101 GAMQ01004936 JAB36915.1 AAGW02052199 AAGW02052200 KPI96716.1 KQ982335 KYQ57538.1 KK853016 KDR12478.1 AZIM01002216 ETE64511.1 PYGN01000274 PSN49552.1
PCG62651.1 KQ460890 KPJ10969.1 KQ459593 KPI96546.1 AGBW02009872 OWR49681.1 PCG62652.1 ODYU01008534 SOQ52219.1 KPI96547.1 AK401394 BAM18016.1 KPJ10970.1 NWSH01002754 PCG67571.1 GDQN01007452 JAT83602.1 ODYU01011594 SOQ57431.1 NWSH01001079 PCG72724.1 NWSH01003298 PCG66594.1 NWSH01006591 PCG63347.1 IACK01084098 LAA80299.1 IACJ01112898 LAA57005.1 ACPB03005685 AAWZ02016749 AAWZ02016750 AAWZ02016751 GEDC01008352 JAS28946.1 NDHI03003384 PNJ71145.1 AHZZ02024143 NEVH01026389 PNF14352.1 AQIB01159356 ABGA01392235 ABGA01392236 ABGA01392237 ABGA01392238 ABGA01392239 ABGA01392240 ABGA01392241 ABGA01392242 AJFE02097058 AK313114 AK300919 AL022724 CH471087 BC007011 AACZ04065924 NBAG03000244 PNI62836.1 JU323455 AFE67211.1 JSUE03030254 JSUE03030255 GEDC01024428 GEDC01024128 GEDC01022375 GEDC01018863 GEDC01018435 GEDC01006808 JAS12870.1 JAS13170.1 JAS14923.1 JAS18435.1 JAS18863.1 JAS30490.1 CM001279 EHH52716.1 ADFV01015550 ADFV01015551 ADFV01015552 CM001256 EHH17956.1 CABD030043317 CABD030043318 CABD030043319 AGTP01048485 AGTP01048486 AGTP01048487 AFYH01049129 AFYH01049130 AFYH01049131 AFYH01049132 AEFK01063730 AEFK01063731 AEFK01063732 AEFK01063733 AEFK01063734 AEFK01063735 AQIA01052761 AQIA01052762 FR904873 CDQ73036.1 AEMK02000051 AANG04001101 GAMQ01004936 JAB36915.1 AAGW02052199 AAGW02052200 KPI96716.1 KQ982335 KYQ57538.1 KK853016 KDR12478.1 AZIM01002216 ETE64511.1 PYGN01000274 PSN49552.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000015103
+ More
UP000001646 UP000233120 UP000028761 UP000235965 UP000029965 UP000233060 UP000001595 UP000240080 UP000005640 UP000002277 UP000006718 UP000252040 UP000009130 UP000001073 UP000001519 UP000233080 UP000233180 UP000233200 UP000005215 UP000245320 UP000008672 UP000007648 UP000233100 UP000002281 UP000087266 UP000193380 UP000008227 UP000011712 UP000233140 UP000189704 UP000221080 UP000233020 UP000001811 UP000008225 UP000075809 UP000027135 UP000245037 UP000233040
UP000001646 UP000233120 UP000028761 UP000235965 UP000029965 UP000233060 UP000001595 UP000240080 UP000005640 UP000002277 UP000006718 UP000252040 UP000009130 UP000001073 UP000001519 UP000233080 UP000233180 UP000233200 UP000005215 UP000245320 UP000008672 UP000007648 UP000233100 UP000002281 UP000087266 UP000193380 UP000008227 UP000011712 UP000233140 UP000189704 UP000221080 UP000233020 UP000001811 UP000008225 UP000075809 UP000027135 UP000245037 UP000233040
Pfam
PF04750 Far-17a_AIG1
Interpro
IPR006838
Far-17a_AIG1
ProteinModelPortal
H9JJI9
H9JJI8
S4P4C5
A0A2A4ISD3
A0A194R0B8
A0A194PSZ5
+ More
A0A212F7H0 A0A2A4ITZ9 A0A2H1WGM3 A0A194PTA5 I4DJC6 A0A194R142 A0A2A4J6I9 A0A1E1W9D6 A0A2H1WWJ5 A0A2A4JMB5 A0A2A4J503 A0A2A4ITN6 A0A2D4I7W0 A0A2D4GB73 T1IBD8 H9GE02 A0A1B6DTE3 A0A2K6D6X9 A0A2K6D6W2 A0A2J8WN06 A0A2I3N3A2 A0A2J7PDG0 A0A0D9R570 A0A2K5MJC8 A0A096NKY2 A0A2K5MJC9 H2PHY1 A0A2R9AV55 Q96IZ2 A0A2R9ASQ7 A0A2I3RF37 Q96IZ2-3 H9EXV2 A0A1D5Q198 A0A1B6CZW9 F7FVQ1 A0A341D0B8 G7P4F3 A0A2I3HSN3 G7MQK8 A0A2K6D6W5 G3S0A2 G3R864 A0A2K5JJ01 A0A2I3ML13 A0A2K6KYY2 A0A2K6RGG7 I3NHT6 A0A2K5MJD1 A0A2U4C5S4 M3XJ79 G3WW37 F7HLH0 A0A2K5WPU0 F6TKL5 A0A1S3MCH5 A0A060X0N9 F1RV85 M3WAX6 A0A2K5Y9N0 A0A1U7TW54 A0A2K5Y9N2 A0A2D0SL67 A0A2K5CIT5 A0A2K5WPV3 F7DN37 G1SDH9 A0A194PTG9 F7EH87 A0A2D0SL66 A0A151XB65 A0A067QU27 V8NRF1 A0A2K5JIW7 A0A2P8YZ85 A0A2K5RAU8 A0A2K6RGJ0 A0A2K6KZ03 F7I3Z6
A0A212F7H0 A0A2A4ITZ9 A0A2H1WGM3 A0A194PTA5 I4DJC6 A0A194R142 A0A2A4J6I9 A0A1E1W9D6 A0A2H1WWJ5 A0A2A4JMB5 A0A2A4J503 A0A2A4ITN6 A0A2D4I7W0 A0A2D4GB73 T1IBD8 H9GE02 A0A1B6DTE3 A0A2K6D6X9 A0A2K6D6W2 A0A2J8WN06 A0A2I3N3A2 A0A2J7PDG0 A0A0D9R570 A0A2K5MJC8 A0A096NKY2 A0A2K5MJC9 H2PHY1 A0A2R9AV55 Q96IZ2 A0A2R9ASQ7 A0A2I3RF37 Q96IZ2-3 H9EXV2 A0A1D5Q198 A0A1B6CZW9 F7FVQ1 A0A341D0B8 G7P4F3 A0A2I3HSN3 G7MQK8 A0A2K6D6W5 G3S0A2 G3R864 A0A2K5JJ01 A0A2I3ML13 A0A2K6KYY2 A0A2K6RGG7 I3NHT6 A0A2K5MJD1 A0A2U4C5S4 M3XJ79 G3WW37 F7HLH0 A0A2K5WPU0 F6TKL5 A0A1S3MCH5 A0A060X0N9 F1RV85 M3WAX6 A0A2K5Y9N0 A0A1U7TW54 A0A2K5Y9N2 A0A2D0SL67 A0A2K5CIT5 A0A2K5WPV3 F7DN37 G1SDH9 A0A194PTG9 F7EH87 A0A2D0SL66 A0A151XB65 A0A067QU27 V8NRF1 A0A2K5JIW7 A0A2P8YZ85 A0A2K5RAU8 A0A2K6RGJ0 A0A2K6KZ03 F7I3Z6
Ontologies
GO
PANTHER
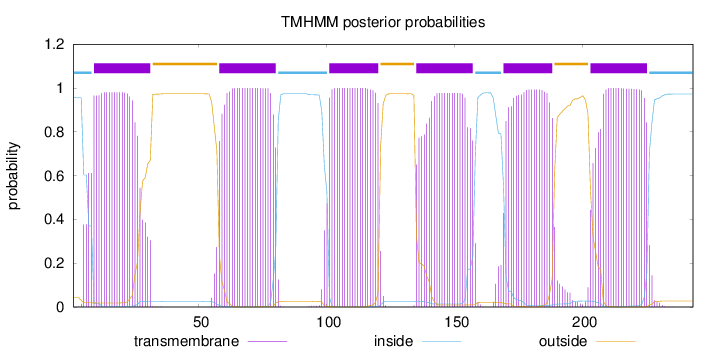
Topology
Subcellular location
Cell membrane
Length:
243
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
127.87844
Exp number, first 60 AAs:
24.1859
Total prob of N-in:
0.95664
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 31
outside
32 - 57
TMhelix
58 - 80
inside
81 - 100
TMhelix
101 - 120
outside
121 - 134
TMhelix
135 - 157
inside
158 - 168
TMhelix
169 - 188
outside
189 - 202
TMhelix
203 - 225
inside
226 - 243
Population Genetic Test Statistics
Pi
225.986555
Theta
143.029386
Tajima's D
1.360296
CLR
0.254041
CSRT
0.748262586870656
Interpretation
Uncertain
