Gene
KWMTBOMO00813
Pre Gene Modal
BGIBMGA009740
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase_CWC27_homolog_isoform_X1_[Bombyx_mori]
Full name
Spliceosome-associated protein CWC27 homolog
+ More
Peptidyl-prolyl cis-trans isomerase
Peptidyl-prolyl cis-trans isomerase
Alternative Name
Probable inactive peptidyl-prolyl cis-trans isomerase CWC27 homolog
Antigen NY-CO-10
Serologically defined colon cancer antigen 10
Antigen NY-CO-10
Serologically defined colon cancer antigen 10
Location in the cell
Nuclear Reliability : 2.395
Sequence
CDS
ATGAGTAATATTTACATACAAGAACCGCCTGCACTAGGAAAGGTTCTTTTGAAGACCACAGCAGGAGAAATTGACATAGAATTATGGACAAAAGAAACCCCGAAAGCATGTCGTAACTTCATACAGCTATGTTTGGAAGGGTACTATAACGGAACAATATTCCATCGTGTGGTGCCAGGTTTCATAGTACAGGGTGGTGATCCAAACGGTGATGGTACTGGAGGAGAATCAGTCTATGGTGCTCCGTTCAAAGATGAGTTCCACTCCCGTCTGCGCTTCAGTCGGCGCGGTCTCGTCGCGATGGCGAACGCCGGCAAAGATGACAACGGCTCCCAGTTCTTCTTCACGCTGGCATCGGCCCCTGAGCTGCAGAACAAGCACACCATCTTCGGGAAGGTCACCGGTGAGACAATATACAATGTACTGAAGCTAGCTGATGGTCTGATCGGTCCGGATGATAGACCGGAACACCCCCACAAGATTCAGAGCACCGAAGTTATTATCAACCCCTTCACTGATATAGTCCCGAGGCTGACGGAGACCAGGGTCGAGGAGTTGCCGAAGAAAAAGAAAAGGGAGCGTCAGGGTGTCAAAAATTTCGGTCTGCTCTCTTTCGGCGAAGAGGCAGAGGAAGACGAAGGCGAAGCCCTGGAGTATAGAGGCAAACCCAAGTCAACACACGATCTTCTAGAAGATCCTAAGCTTAGCAAAAAGACTGCTGCAGAGCTGGAGACGCAAGTCGATAAAAAGACAAACGTGGAAATGGCTAAAGAAATTGAAGAGAAAAGACTGGAGACAGAGGCCACGGTCAGCAGCATACGAGACAAATTGAGTTCGAAGAGGGAGAGCAGCGTCGACAGGGAGACAAAGAGACAGAAACAGGAATGCGAGAGCGAGTCGGACGGAGAATATTACATAGGGAAAGACAGAGACATCGAGAGGAAGAAAGAGAGGTGA
Protein
MSNIYIQEPPALGKVLLKTTAGEIDIELWTKETPKACRNFIQLCLEGYYNGTIFHRVVPGFIVQGGDPNGDGTGGESVYGAPFKDEFHSRLRFSRRGLVAMANAGKDDNGSQFFFTLASAPELQNKHTIFGKVTGETIYNVLKLADGLIGPDDRPEHPHKIQSTEVIINPFTDIVPRLTETRVEELPKKKKRERQGVKNFGLLSFGEEAEEDEGEALEYRGKPKSTHDLLEDPKLSKKTAAELETQVDKKTNVEMAKEIEEKRLETEATVSSIRDKLSSKRESSVDRETKRQKQECESESDGEYYIGKDRDIERKKER
Summary
Description
As part of the spliceosome, plays a role in pre-mRNA splicing. Probable inactive PPIase with no peptidyl-prolyl cis-trans isomerase activity.
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
As part of the spliceosome, plays a role in pre-mRNA splicing (PubMed:29360106). Probable inactive PPIase with no peptidyl-prolyl cis-trans isomerase activity (PubMed:20676357).
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
As part of the spliceosome, plays a role in pre-mRNA splicing (PubMed:29360106). Probable inactive PPIase with no peptidyl-prolyl cis-trans isomerase activity (PubMed:20676357).
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Subunit
Part of the activated spliceosome B/catalytic step 1 spliceosome, one of the forms of the spliceosome which has a well-formed active site but still cannot catalyze the branching reaction and is composed at least of 52 proteins, the U2, U5 and U6 snRNAs and the pre-mRNA. Recruited during early steps of activated spliceosome B maturation, it is probably one of the first proteins released from this complex as he matures to the spliceosome C complex.
Similarity
Belongs to the cyclophilin-type PPIase family.
Keywords
Coiled coil
Nucleus
Phosphoprotein
Complete proteome
Reference proteome
Acetylation
3D-structure
Alternative splicing
Disease mutation
Dwarfism
Glycoprotein
Mental retardation
Polymorphism
Retinitis pigmentosa
Feature
chain Spliceosome-associated protein CWC27 homolog
splice variant In isoform 2.
sequence variant In RPSKA.
splice variant In isoform 2.
sequence variant In RPSKA.
Uniprot
H9JJN9
A0A2H1VNJ8
A0A2A4ITM9
A0A194PTA1
A0A212F4B1
A0A2P8Y6C9
+ More
E2BNU1 A0A0J7P4A2 A0A087ZQ64 A0A0M9A2J1 A0A1B6MUA0 A0A194R138 A0A3L8DGA2 A0A293M4M3 A0A2P6L3E8 A0A158P3K5 A0A1Z5LH07 A0A1B6GDT2 A0A1B0CMB8 A0A310SE84 A0A0L7R4G3 A0A1B6HYF7 Q9VTN7 H0YX50 A0A218V9U6 B4HEZ1 B4N735 B4KUU2 A0A182JAS3 A0A1I9WLJ3 B4QQN0 B3NH08 A0A340WF93 A0A1L8DTX9 A0A026WXY4 G3VDF3 A0A210R5L4 Q6GLX7 A0A1W4VTB7 E1ZX62 B4LCS8 B4PFP5 A0A151IH65 A0A3B4AWQ6 V8NPM4 F1QRY1 Q7ZW86 A0A195F1C5 A0A2A3EJ52 A0A2I0M1A8 A0A182UTV2 A0A151IV62 A0A3B0J4I7 A0A087TA95 A0A0A1XA14 A0A0Q3QSB0 U3K554 A0A1U7TWG8 A0A2J7PD97 A0A061IK06 W5JG65 A0A0K8WDJ6 Q3TKY6 A0A2K6EGX1 G3HXI0 E9J8R4 A0A2D0SN47 Q29DY0 B4H480 G1KRD8 A0A0N8ET94 A0A0M3QWB3 U3KMX5 A0A2D0SPC0 A0A2D0SNQ7 U4UYM0 A0A1S3AFQ7 A0A2K6EGV9 A0A226N4W8 A0A151MXD7 I3LW47 Q6UX04-2 J3S472 B3M4C7 N6UHJ3 A0A2Y9SD63 A0A0G2JXR7 A0A1S3AFG2 A0A250Y1H9 A0A2U3XM69 F1SKU1 A0A2K5BUZ5 A0A2Y9I1G9 A0A182WIY6 A0A2Y9Q9W9 A0A2K6T450 F6XPF6 Q6UX04 A0A2I2ZMX7 E2R440
E2BNU1 A0A0J7P4A2 A0A087ZQ64 A0A0M9A2J1 A0A1B6MUA0 A0A194R138 A0A3L8DGA2 A0A293M4M3 A0A2P6L3E8 A0A158P3K5 A0A1Z5LH07 A0A1B6GDT2 A0A1B0CMB8 A0A310SE84 A0A0L7R4G3 A0A1B6HYF7 Q9VTN7 H0YX50 A0A218V9U6 B4HEZ1 B4N735 B4KUU2 A0A182JAS3 A0A1I9WLJ3 B4QQN0 B3NH08 A0A340WF93 A0A1L8DTX9 A0A026WXY4 G3VDF3 A0A210R5L4 Q6GLX7 A0A1W4VTB7 E1ZX62 B4LCS8 B4PFP5 A0A151IH65 A0A3B4AWQ6 V8NPM4 F1QRY1 Q7ZW86 A0A195F1C5 A0A2A3EJ52 A0A2I0M1A8 A0A182UTV2 A0A151IV62 A0A3B0J4I7 A0A087TA95 A0A0A1XA14 A0A0Q3QSB0 U3K554 A0A1U7TWG8 A0A2J7PD97 A0A061IK06 W5JG65 A0A0K8WDJ6 Q3TKY6 A0A2K6EGX1 G3HXI0 E9J8R4 A0A2D0SN47 Q29DY0 B4H480 G1KRD8 A0A0N8ET94 A0A0M3QWB3 U3KMX5 A0A2D0SPC0 A0A2D0SNQ7 U4UYM0 A0A1S3AFQ7 A0A2K6EGV9 A0A226N4W8 A0A151MXD7 I3LW47 Q6UX04-2 J3S472 B3M4C7 N6UHJ3 A0A2Y9SD63 A0A0G2JXR7 A0A1S3AFG2 A0A250Y1H9 A0A2U3XM69 F1SKU1 A0A2K5BUZ5 A0A2Y9I1G9 A0A182WIY6 A0A2Y9Q9W9 A0A2K6T450 F6XPF6 Q6UX04 A0A2I2ZMX7 E2R440
EC Number
5.2.1.8
Pubmed
19121390
26354079
22118469
29403074
20798317
30249741
+ More
21347285 28528879 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20360741 17994087 27538518 22936249 24508170 21709235 28812685 17550304 25463417 24297900 23594743 23371554 25830018 23929341 20920257 23761445 16141072 15489334 21183079 28285769 21804562 21282665 15632085 21993624 23537049 22293439 9610721 12975309 16335952 19413330 21269460 23186163 20676357 25478830 29360106 23025625 15057822 22673903 28087693 30723633 17495919 22398555 16341006
21347285 28528879 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20360741 17994087 27538518 22936249 24508170 21709235 28812685 17550304 25463417 24297900 23594743 23371554 25830018 23929341 20920257 23761445 16141072 15489334 21183079 28285769 21804562 21282665 15632085 21993624 23537049 22293439 9610721 12975309 16335952 19413330 21269460 23186163 20676357 25478830 29360106 23025625 15057822 22673903 28087693 30723633 17495919 22398555 16341006
EMBL
BABH01017463
BABH01017464
BABH01017465
BABH01017466
ODYU01003512
SOQ42393.1
+ More
NWSH01007247 PCG63029.1 KQ459593 KPI96542.1 AGBW02010396 OWR48576.1 PYGN01000874 PSN39805.1 GL449507 EFN82624.1 LBMM01000143 KMR04764.1 KQ435756 KOX75897.1 GEBQ01000451 JAT39526.1 KQ460890 KPJ10965.1 QOIP01000008 RLU19222.1 GFWV01010527 MAA35256.1 MWRG01002072 PRD33101.1 ADTU01008167 GFJQ02000284 JAW06686.1 GECZ01009316 JAS60453.1 AJWK01018556 KQ767768 OAD53306.1 KQ414658 KOC65646.1 GECU01027988 JAS79718.1 AE014296 AY070988 AAF50009.1 AAL48610.1 ABQF01033307 ABQF01033308 ABQF01033309 MUZQ01000025 OWK62530.1 CH480815 EDW41160.1 CH964168 EDW80174.1 CH933809 EDW19348.1 KU932377 APA34013.1 CM000363 CM002912 EDX10132.1 KMY99073.1 CH954178 EDV51465.1 GFDF01004171 JAV09913.1 KK107064 EZA60902.1 AEFK01050328 AEFK01050329 AEFK01050330 AEFK01050331 AEFK01050332 AEFK01050333 AEFK01050334 AEFK01050335 AEFK01050336 AEFK01050337 NEDP02000216 OWF56333.1 BC074317 GL435030 EFN74152.1 CH940647 EDW70970.1 CM000159 EDW94194.1 KQ977636 KYN01148.1 AZIM01002721 ETE63472.1 BX640403 BC049533 KQ981880 KYN33904.1 KZ288229 PBC31718.1 AKCR02000050 PKK23464.1 KQ980924 KYN11417.1 OUUW01000002 SPP76754.1 KK114261 KFM62034.1 GBXI01006350 JAD07942.1 LMAW01002889 KQK76017.1 AGTO01020873 NEVH01026392 PNF14301.1 KE668376 ERE83944.1 ADMH02001258 ETN63362.1 GDHF01003101 JAI49213.1 AK014025 AK075902 AK077779 AK166770 BC025437 JH000876 EGW03509.1 GL769036 EFZ10801.1 CH379070 EAL30283.2 CH479208 EDW31195.1 GEBF01005011 JAN98621.1 CP012525 ALC43849.1 AAGW02031073 AAGW02031074 AAGW02031075 AAGW02031076 AAGW02031077 AAGW02031078 KB632430 ERL95491.1 MCFN01000211 OXB62522.1 AKHW03004724 KYO29177.1 AGTP01014042 AGTP01014043 AGTP01014044 AGTP01014045 AGTP01014046 AF039692 AF039693 AY358569 CH471137 BC012117 JU174261 GBEX01000833 AFJ49787.1 JAI13727.1 CH902618 EDV39397.1 APGK01034947 KB740923 ENN78097.1 AABR07007909 AABR07007910 AABR07007911 AABR07007912 GFFV01002484 JAV37461.1 AEMK02000102 DQIR01257324 HDC12802.1 CABD030104574 CABD030104575 CABD030104576 AAEX03001470 AAEX03001471
NWSH01007247 PCG63029.1 KQ459593 KPI96542.1 AGBW02010396 OWR48576.1 PYGN01000874 PSN39805.1 GL449507 EFN82624.1 LBMM01000143 KMR04764.1 KQ435756 KOX75897.1 GEBQ01000451 JAT39526.1 KQ460890 KPJ10965.1 QOIP01000008 RLU19222.1 GFWV01010527 MAA35256.1 MWRG01002072 PRD33101.1 ADTU01008167 GFJQ02000284 JAW06686.1 GECZ01009316 JAS60453.1 AJWK01018556 KQ767768 OAD53306.1 KQ414658 KOC65646.1 GECU01027988 JAS79718.1 AE014296 AY070988 AAF50009.1 AAL48610.1 ABQF01033307 ABQF01033308 ABQF01033309 MUZQ01000025 OWK62530.1 CH480815 EDW41160.1 CH964168 EDW80174.1 CH933809 EDW19348.1 KU932377 APA34013.1 CM000363 CM002912 EDX10132.1 KMY99073.1 CH954178 EDV51465.1 GFDF01004171 JAV09913.1 KK107064 EZA60902.1 AEFK01050328 AEFK01050329 AEFK01050330 AEFK01050331 AEFK01050332 AEFK01050333 AEFK01050334 AEFK01050335 AEFK01050336 AEFK01050337 NEDP02000216 OWF56333.1 BC074317 GL435030 EFN74152.1 CH940647 EDW70970.1 CM000159 EDW94194.1 KQ977636 KYN01148.1 AZIM01002721 ETE63472.1 BX640403 BC049533 KQ981880 KYN33904.1 KZ288229 PBC31718.1 AKCR02000050 PKK23464.1 KQ980924 KYN11417.1 OUUW01000002 SPP76754.1 KK114261 KFM62034.1 GBXI01006350 JAD07942.1 LMAW01002889 KQK76017.1 AGTO01020873 NEVH01026392 PNF14301.1 KE668376 ERE83944.1 ADMH02001258 ETN63362.1 GDHF01003101 JAI49213.1 AK014025 AK075902 AK077779 AK166770 BC025437 JH000876 EGW03509.1 GL769036 EFZ10801.1 CH379070 EAL30283.2 CH479208 EDW31195.1 GEBF01005011 JAN98621.1 CP012525 ALC43849.1 AAGW02031073 AAGW02031074 AAGW02031075 AAGW02031076 AAGW02031077 AAGW02031078 KB632430 ERL95491.1 MCFN01000211 OXB62522.1 AKHW03004724 KYO29177.1 AGTP01014042 AGTP01014043 AGTP01014044 AGTP01014045 AGTP01014046 AF039692 AF039693 AY358569 CH471137 BC012117 JU174261 GBEX01000833 AFJ49787.1 JAI13727.1 CH902618 EDV39397.1 APGK01034947 KB740923 ENN78097.1 AABR07007909 AABR07007910 AABR07007911 AABR07007912 GFFV01002484 JAV37461.1 AEMK02000102 DQIR01257324 HDC12802.1 CABD030104574 CABD030104575 CABD030104576 AAEX03001470 AAEX03001471
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000245037
UP000008237
+ More
UP000036403 UP000005203 UP000053105 UP000053240 UP000279307 UP000005205 UP000092461 UP000053825 UP000000803 UP000007754 UP000197619 UP000001292 UP000007798 UP000009192 UP000075880 UP000000304 UP000008711 UP000265300 UP000053097 UP000007648 UP000242188 UP000192221 UP000000311 UP000008792 UP000002282 UP000078542 UP000261520 UP000000437 UP000078541 UP000242457 UP000053872 UP000075903 UP000078492 UP000268350 UP000054359 UP000051836 UP000016665 UP000189704 UP000235965 UP000030759 UP000000673 UP000000589 UP000233160 UP000001075 UP000221080 UP000001819 UP000008744 UP000001646 UP000092553 UP000001811 UP000030742 UP000079721 UP000198323 UP000050525 UP000005215 UP000005640 UP000007801 UP000019118 UP000248484 UP000002494 UP000245341 UP000008227 UP000233020 UP000248481 UP000075920 UP000248483 UP000233220 UP000002280 UP000001519 UP000002254
UP000036403 UP000005203 UP000053105 UP000053240 UP000279307 UP000005205 UP000092461 UP000053825 UP000000803 UP000007754 UP000197619 UP000001292 UP000007798 UP000009192 UP000075880 UP000000304 UP000008711 UP000265300 UP000053097 UP000007648 UP000242188 UP000192221 UP000000311 UP000008792 UP000002282 UP000078542 UP000261520 UP000000437 UP000078541 UP000242457 UP000053872 UP000075903 UP000078492 UP000268350 UP000054359 UP000051836 UP000016665 UP000189704 UP000235965 UP000030759 UP000000673 UP000000589 UP000233160 UP000001075 UP000221080 UP000001819 UP000008744 UP000001646 UP000092553 UP000001811 UP000030742 UP000079721 UP000198323 UP000050525 UP000005215 UP000005640 UP000007801 UP000019118 UP000248484 UP000002494 UP000245341 UP000008227 UP000233020 UP000248481 UP000075920 UP000248483 UP000233220 UP000002280 UP000001519 UP000002254
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JJN9
A0A2H1VNJ8
A0A2A4ITM9
A0A194PTA1
A0A212F4B1
A0A2P8Y6C9
+ More
E2BNU1 A0A0J7P4A2 A0A087ZQ64 A0A0M9A2J1 A0A1B6MUA0 A0A194R138 A0A3L8DGA2 A0A293M4M3 A0A2P6L3E8 A0A158P3K5 A0A1Z5LH07 A0A1B6GDT2 A0A1B0CMB8 A0A310SE84 A0A0L7R4G3 A0A1B6HYF7 Q9VTN7 H0YX50 A0A218V9U6 B4HEZ1 B4N735 B4KUU2 A0A182JAS3 A0A1I9WLJ3 B4QQN0 B3NH08 A0A340WF93 A0A1L8DTX9 A0A026WXY4 G3VDF3 A0A210R5L4 Q6GLX7 A0A1W4VTB7 E1ZX62 B4LCS8 B4PFP5 A0A151IH65 A0A3B4AWQ6 V8NPM4 F1QRY1 Q7ZW86 A0A195F1C5 A0A2A3EJ52 A0A2I0M1A8 A0A182UTV2 A0A151IV62 A0A3B0J4I7 A0A087TA95 A0A0A1XA14 A0A0Q3QSB0 U3K554 A0A1U7TWG8 A0A2J7PD97 A0A061IK06 W5JG65 A0A0K8WDJ6 Q3TKY6 A0A2K6EGX1 G3HXI0 E9J8R4 A0A2D0SN47 Q29DY0 B4H480 G1KRD8 A0A0N8ET94 A0A0M3QWB3 U3KMX5 A0A2D0SPC0 A0A2D0SNQ7 U4UYM0 A0A1S3AFQ7 A0A2K6EGV9 A0A226N4W8 A0A151MXD7 I3LW47 Q6UX04-2 J3S472 B3M4C7 N6UHJ3 A0A2Y9SD63 A0A0G2JXR7 A0A1S3AFG2 A0A250Y1H9 A0A2U3XM69 F1SKU1 A0A2K5BUZ5 A0A2Y9I1G9 A0A182WIY6 A0A2Y9Q9W9 A0A2K6T450 F6XPF6 Q6UX04 A0A2I2ZMX7 E2R440
E2BNU1 A0A0J7P4A2 A0A087ZQ64 A0A0M9A2J1 A0A1B6MUA0 A0A194R138 A0A3L8DGA2 A0A293M4M3 A0A2P6L3E8 A0A158P3K5 A0A1Z5LH07 A0A1B6GDT2 A0A1B0CMB8 A0A310SE84 A0A0L7R4G3 A0A1B6HYF7 Q9VTN7 H0YX50 A0A218V9U6 B4HEZ1 B4N735 B4KUU2 A0A182JAS3 A0A1I9WLJ3 B4QQN0 B3NH08 A0A340WF93 A0A1L8DTX9 A0A026WXY4 G3VDF3 A0A210R5L4 Q6GLX7 A0A1W4VTB7 E1ZX62 B4LCS8 B4PFP5 A0A151IH65 A0A3B4AWQ6 V8NPM4 F1QRY1 Q7ZW86 A0A195F1C5 A0A2A3EJ52 A0A2I0M1A8 A0A182UTV2 A0A151IV62 A0A3B0J4I7 A0A087TA95 A0A0A1XA14 A0A0Q3QSB0 U3K554 A0A1U7TWG8 A0A2J7PD97 A0A061IK06 W5JG65 A0A0K8WDJ6 Q3TKY6 A0A2K6EGX1 G3HXI0 E9J8R4 A0A2D0SN47 Q29DY0 B4H480 G1KRD8 A0A0N8ET94 A0A0M3QWB3 U3KMX5 A0A2D0SPC0 A0A2D0SNQ7 U4UYM0 A0A1S3AFQ7 A0A2K6EGV9 A0A226N4W8 A0A151MXD7 I3LW47 Q6UX04-2 J3S472 B3M4C7 N6UHJ3 A0A2Y9SD63 A0A0G2JXR7 A0A1S3AFG2 A0A250Y1H9 A0A2U3XM69 F1SKU1 A0A2K5BUZ5 A0A2Y9I1G9 A0A182WIY6 A0A2Y9Q9W9 A0A2K6T450 F6XPF6 Q6UX04 A0A2I2ZMX7 E2R440
PDB
4R3E
E-value=1.97709e-75,
Score=717
Ontologies
GO
Topology
Subcellular location
Nucleus
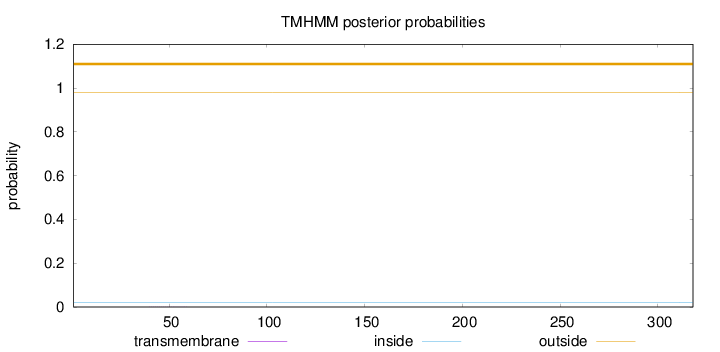
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00446
Exp number, first 60 AAs:
0.0041
Total prob of N-in:
0.02158
outside
1 - 318
Population Genetic Test Statistics
Pi
334.814359
Theta
167.070109
Tajima's D
3.455686
CLR
0.029179
CSRT
0.992700364981751
Interpretation
Uncertain
