Gene
KWMTBOMO00811
Pre Gene Modal
BGIBMGA009693
Annotation
PREDICTED:_angiotensin-converting_enzyme-like_[Bombyx_mori]
Full name
Angiotensin-converting enzyme
Alternative Name
Dipeptidyl carboxypeptidase I
Kininase II
Kininase II
Location in the cell
Mitochondrial Reliability : 1.449 Nuclear Reliability : 1.056
Sequence
CDS
ATGGCAGCTACGGAAAACCTTGGAAGCAACGATGTTTTGTGCTTCAAAGGAGAAGATGATTTTGAAAAATTAATGAGTAGATCAAAGAATGAACGTGTTTTGAAATGGACTTGGATGGCTTGGCGGGAGGCGGTGTGTCGGACAATGGGAGCGCCTTATAAAGAACTGGTTGAAATTGAAAATGCAGCTGCCAGAAGGAACGGCTACAAAGACATCGGCGAGTCCTGGAGAGAGGAACAAGAAATTCCAAATTTAAAAAGTTTTTGTCGTCGCCTATACAAAAGCATAAGACCTTTATACAGGACGCTGCATGGTATCACCAGGTTCTTCCTTAGAAGGCATTACGGGGCAATTGTAGAAGAGACCGGGCCAATACCGGCTCATTTATTGGGAAATCTCTGGTCACAGAATTGGGAGCCGTTAACGGATCTAATAATAAAAAACACGTTGAATTTAGACGAACGCATACGAAGATCTAACTGGACCGTTCTGCATATGGTGAGGATAAAAAAACTAGAAGCTGATTAA
Protein
MAATENLGSNDVLCFKGEDDFEKLMSRSKNERVLKWTWMAWREAVCRTMGAPYKELVEIENAAARRNGYKDIGESWREEQEIPNLKSFCRRLYKSIRPLYRTLHGITRFFLRRHYGAIVEETGPIPAHLLGNLWSQNWEPLTDLIIKNTLNLDERIRRSNWTVLHMVRIKKLEAD
Summary
Description
Converts angiotensin I to angiotensin II by release of the terminal His-Leu, this results in an increase of the vasoconstrictor activity of angiotensin. Also able to inactivate bradykinin, a potent vasodilator. Has also a glycosidase activity which releases GPI-anchored proteins from the membrane by cleaving the mannose linkage in the GPI moiety (By similarity).
Catalytic Activity
Release of a C-terminal dipeptide, oligopeptide-|-Xaa-Yaa, when Xaa is not Pro, and Yaa is neither Asp nor Glu. Thus, conversion of angiotensin I to angiotensin II, with increase in vasoconstrictor activity, but no action on angiotensin II.
Cofactor
Zn(2+)
chloride
chloride
Biophysicochemical Properties
2.51 mM for Hip-His-Leu
Miscellaneous
Inhibitors of ACE are commonly used to treat hypertension and some types of renal and cardiac dysfunction.
The glycosidase activity probably uses different active site residues than the metalloprotease activity.
The glycosidase activity probably uses different active site residues than the metalloprotease activity.
Similarity
Belongs to the peptidase M2 family.
Keywords
3D-structure
Alternative splicing
Carboxypeptidase
Cell membrane
Complete proteome
Cytoplasm
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Membrane
Metal-binding
Metalloprotease
Phosphoprotein
Polymorphism
Protease
Reference proteome
Repeat
Secreted
Signal
Transmembrane
Transmembrane helix
Zinc
Feature
chain Angiotensin-converting enzyme
propeptide Removed in soluble form
splice variant In isoform 4.
sequence variant In dbSNP:rs13306087.
propeptide Removed in soluble form
splice variant In isoform 4.
sequence variant In dbSNP:rs13306087.
Uniprot
H9JJJ2
A0A2A4J3I3
A0A2H1VAI7
A0A212FC18
A0A194PSY9
A0A0L7LGJ2
+ More
E2C639 A0A195CS67 A0A067R743 E9JD70 E2ARV1 A0A2J7R0L7 A0A026WAE2 A0A3L8DWH2 Q16KU3 A0A1S4FXA3 W5JAF0 A0A182FFW1 E0VHF1 A0A182TY41 A0A182P3K9 A0A182RFM6 A0A182UVU7 A0A1B0GPC4 A0A182WIQ8 A0A182X6D2 D6WRC2 T1HGC9 Q7QJR1 A0A182KRS0 A0A182I3N8 A0A182QPM5 A0A2K6G9P4 A0A2K6G9P9 A0A182LXR3 A0A182NHM3 A0A2K6G9P1 J3KTB8 A0A1S3GIY4 A0A1D5QY90 L5KPC7 A0A384CCH2 Q3HTU3 A0A2K5CQL4 A0A2R8PBI1 F6R5R2 A0A2K6BQE6 A0A2K5VX29 A0A1B0CRV9 A0A2Y9G5A0 A0A2I3N885 A0A2K6P2V7 A0A2U3YBQ9 A0A182XZ84 G1TYT4 A0A2K6K394 A0A2K5I5V9 A0A2K5XHN5 A0A2I2Y2C2 A0A2R8ZYP4 H2QDM6 F6X3S4 I6L9B3 A0A2K6TDD2 A0A2R8N4Y1 F6QVU8 A0A2K5CQL7 A0A2U3YC18 A0A2K6K3B6 A0A2J8UZC8 A0A2U3YBR9 A0A2K6TDC7 F7F5C6 A0A2U4C3T3 A0A2K5I630 U5L195 P12821-3 A0A2K5CQI8 A0A2K5XHN8 Q9GLN7-2 A0A2R8ZYP1 A0A2U3WBW4 A0A2Y9G4W0 G3RZY8 A0A2K5PZN4 D3DU13 A0A2U3WBV7 A0A2K5VWR1 A0A2K6BQJ6 P12821-4 A0A2K6K3D3 A0A2K6P2X8 A0A096P6D3 A0A2R8ZYN2 A0A2I3SKE0 A0A2K5I5V3 A0A2I2ZRS8 A0A2K5VWS6 A0A1D5QHM2
E2C639 A0A195CS67 A0A067R743 E9JD70 E2ARV1 A0A2J7R0L7 A0A026WAE2 A0A3L8DWH2 Q16KU3 A0A1S4FXA3 W5JAF0 A0A182FFW1 E0VHF1 A0A182TY41 A0A182P3K9 A0A182RFM6 A0A182UVU7 A0A1B0GPC4 A0A182WIQ8 A0A182X6D2 D6WRC2 T1HGC9 Q7QJR1 A0A182KRS0 A0A182I3N8 A0A182QPM5 A0A2K6G9P4 A0A2K6G9P9 A0A182LXR3 A0A182NHM3 A0A2K6G9P1 J3KTB8 A0A1S3GIY4 A0A1D5QY90 L5KPC7 A0A384CCH2 Q3HTU3 A0A2K5CQL4 A0A2R8PBI1 F6R5R2 A0A2K6BQE6 A0A2K5VX29 A0A1B0CRV9 A0A2Y9G5A0 A0A2I3N885 A0A2K6P2V7 A0A2U3YBQ9 A0A182XZ84 G1TYT4 A0A2K6K394 A0A2K5I5V9 A0A2K5XHN5 A0A2I2Y2C2 A0A2R8ZYP4 H2QDM6 F6X3S4 I6L9B3 A0A2K6TDD2 A0A2R8N4Y1 F6QVU8 A0A2K5CQL7 A0A2U3YC18 A0A2K6K3B6 A0A2J8UZC8 A0A2U3YBR9 A0A2K6TDC7 F7F5C6 A0A2U4C3T3 A0A2K5I630 U5L195 P12821-3 A0A2K5CQI8 A0A2K5XHN8 Q9GLN7-2 A0A2R8ZYP1 A0A2U3WBW4 A0A2Y9G4W0 G3RZY8 A0A2K5PZN4 D3DU13 A0A2U3WBV7 A0A2K5VWR1 A0A2K6BQJ6 P12821-4 A0A2K6K3D3 A0A2K6P2X8 A0A096P6D3 A0A2R8ZYN2 A0A2I3SKE0 A0A2K5I5V3 A0A2I2ZRS8 A0A2K5VWS6 A0A1D5QHM2
EC Number
3.4.-.-
Pubmed
19121390
22118469
26354079
26227816
20798317
24845553
+ More
21282665 24508170 30249741 17510324 20920257 23761445 20566863 18362917 19820115 12364791 14747013 17210077 20966253 16625196 17431167 23258410 25362486 25244985 21993624 22398555 22722832 16136131 15489334 2849100 2547653 2554286 10319862 14702039 2558109 9642152 1649623 8755737 9013598 10769174 10969042 10924499 11076943 12386153 12459472 12542396 15151696 15671045 16335952 19159218 21269460 12540854 15236580 16476442 10099885 10391210 11551873 14694062 15534175 15277638 16116425 25787250 11013071 11181995
21282665 24508170 30249741 17510324 20920257 23761445 20566863 18362917 19820115 12364791 14747013 17210077 20966253 16625196 17431167 23258410 25362486 25244985 21993624 22398555 22722832 16136131 15489334 2849100 2547653 2554286 10319862 14702039 2558109 9642152 1649623 8755737 9013598 10769174 10969042 10924499 11076943 12386153 12459472 12542396 15151696 15671045 16335952 19159218 21269460 12540854 15236580 16476442 10099885 10391210 11551873 14694062 15534175 15277638 16116425 25787250 11013071 11181995
EMBL
BABH01017456
BABH01017457
BABH01017458
BABH01017459
BABH01017460
NWSH01003593
+ More
PCG66074.1 ODYU01001260 SOQ37224.1 AGBW02009238 OWR51282.1 KQ459593 KPI96541.1 JTDY01001279 KOB74311.1 GL452916 EFN76582.1 KQ977329 KYN03578.1 KK852657 KDR19225.1 GL771866 EFZ09138.1 GL442175 EFN63834.1 NEVH01008227 PNF34382.1 KK107295 EZA53025.1 QOIP01000003 RLU24850.1 CH477938 EAT34915.1 ADMH02001692 ETN61407.1 DS235169 EEB12807.1 AJVK01033349 KQ971351 EFA06546.2 ACPB03020413 ACPB03020414 AAAB01008807 EAA03963.4 APCN01000215 AXCN02000593 AXCM01000215 AC113554 JSUE03017170 JSUE03017171 KB030661 ELK12483.1 DQ195091 ABA40748.1 AQIA01028261 AJWK01025349 AHZZ02010522 AHZZ02010523 AAGW02039959 CABD030036630 CABD030036631 AJFE02035554 AJFE02035555 AJFE02035556 AJFE02035557 AACZ04070149 AACZ04070150 NBAG03000001 PNJ01391.1 BC036375 AAH36375.1 NDHI03003438 PNJ50625.1 KC153308 AGV55421.1 J04144 M26657 X16295 AF118569 AY436326 AK301988 EU332840 AB208971 AF193486 AF193462 AF193464 AF193463 AF193465 AF193467 AF193469 AF193471 AF193473 AF193482 AF193481 AF193480 AF193479 AF193478 AF193477 AF193476 AF193475 AF193474 AF193485 AF193484 AF193483 AF193472 AF193470 AF193468 AF193466 CH471109 EAW94321.1
PCG66074.1 ODYU01001260 SOQ37224.1 AGBW02009238 OWR51282.1 KQ459593 KPI96541.1 JTDY01001279 KOB74311.1 GL452916 EFN76582.1 KQ977329 KYN03578.1 KK852657 KDR19225.1 GL771866 EFZ09138.1 GL442175 EFN63834.1 NEVH01008227 PNF34382.1 KK107295 EZA53025.1 QOIP01000003 RLU24850.1 CH477938 EAT34915.1 ADMH02001692 ETN61407.1 DS235169 EEB12807.1 AJVK01033349 KQ971351 EFA06546.2 ACPB03020413 ACPB03020414 AAAB01008807 EAA03963.4 APCN01000215 AXCN02000593 AXCM01000215 AC113554 JSUE03017170 JSUE03017171 KB030661 ELK12483.1 DQ195091 ABA40748.1 AQIA01028261 AJWK01025349 AHZZ02010522 AHZZ02010523 AAGW02039959 CABD030036630 CABD030036631 AJFE02035554 AJFE02035555 AJFE02035556 AJFE02035557 AACZ04070149 AACZ04070150 NBAG03000001 PNJ01391.1 BC036375 AAH36375.1 NDHI03003438 PNJ50625.1 KC153308 AGV55421.1 J04144 M26657 X16295 AF118569 AY436326 AK301988 EU332840 AB208971 AF193486 AF193462 AF193464 AF193463 AF193465 AF193467 AF193469 AF193471 AF193473 AF193482 AF193481 AF193480 AF193479 AF193478 AF193477 AF193476 AF193475 AF193474 AF193485 AF193484 AF193483 AF193472 AF193470 AF193468 AF193466 CH471109 EAW94321.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000008237
+ More
UP000078542 UP000027135 UP000000311 UP000235965 UP000053097 UP000279307 UP000008820 UP000000673 UP000069272 UP000009046 UP000075902 UP000075885 UP000075900 UP000075903 UP000092462 UP000075920 UP000076407 UP000007266 UP000015103 UP000007062 UP000075882 UP000075840 UP000075886 UP000233160 UP000075883 UP000075884 UP000005640 UP000081671 UP000006718 UP000010552 UP000261680 UP000233020 UP000008225 UP000233120 UP000233100 UP000092461 UP000248481 UP000028761 UP000233200 UP000245341 UP000076408 UP000001811 UP000233180 UP000233080 UP000233140 UP000001519 UP000240080 UP000002277 UP000233220 UP000245320 UP000245340 UP000233040
UP000078542 UP000027135 UP000000311 UP000235965 UP000053097 UP000279307 UP000008820 UP000000673 UP000069272 UP000009046 UP000075902 UP000075885 UP000075900 UP000075903 UP000092462 UP000075920 UP000076407 UP000007266 UP000015103 UP000007062 UP000075882 UP000075840 UP000075886 UP000233160 UP000075883 UP000075884 UP000005640 UP000081671 UP000006718 UP000010552 UP000261680 UP000233020 UP000008225 UP000233120 UP000233100 UP000092461 UP000248481 UP000028761 UP000233200 UP000245341 UP000076408 UP000001811 UP000233180 UP000233080 UP000233140 UP000001519 UP000240080 UP000002277 UP000233220 UP000245320 UP000245340 UP000233040
SUPFAM
SSF81427
SSF81427
Gene 3D
ProteinModelPortal
H9JJJ2
A0A2A4J3I3
A0A2H1VAI7
A0A212FC18
A0A194PSY9
A0A0L7LGJ2
+ More
E2C639 A0A195CS67 A0A067R743 E9JD70 E2ARV1 A0A2J7R0L7 A0A026WAE2 A0A3L8DWH2 Q16KU3 A0A1S4FXA3 W5JAF0 A0A182FFW1 E0VHF1 A0A182TY41 A0A182P3K9 A0A182RFM6 A0A182UVU7 A0A1B0GPC4 A0A182WIQ8 A0A182X6D2 D6WRC2 T1HGC9 Q7QJR1 A0A182KRS0 A0A182I3N8 A0A182QPM5 A0A2K6G9P4 A0A2K6G9P9 A0A182LXR3 A0A182NHM3 A0A2K6G9P1 J3KTB8 A0A1S3GIY4 A0A1D5QY90 L5KPC7 A0A384CCH2 Q3HTU3 A0A2K5CQL4 A0A2R8PBI1 F6R5R2 A0A2K6BQE6 A0A2K5VX29 A0A1B0CRV9 A0A2Y9G5A0 A0A2I3N885 A0A2K6P2V7 A0A2U3YBQ9 A0A182XZ84 G1TYT4 A0A2K6K394 A0A2K5I5V9 A0A2K5XHN5 A0A2I2Y2C2 A0A2R8ZYP4 H2QDM6 F6X3S4 I6L9B3 A0A2K6TDD2 A0A2R8N4Y1 F6QVU8 A0A2K5CQL7 A0A2U3YC18 A0A2K6K3B6 A0A2J8UZC8 A0A2U3YBR9 A0A2K6TDC7 F7F5C6 A0A2U4C3T3 A0A2K5I630 U5L195 P12821-3 A0A2K5CQI8 A0A2K5XHN8 Q9GLN7-2 A0A2R8ZYP1 A0A2U3WBW4 A0A2Y9G4W0 G3RZY8 A0A2K5PZN4 D3DU13 A0A2U3WBV7 A0A2K5VWR1 A0A2K6BQJ6 P12821-4 A0A2K6K3D3 A0A2K6P2X8 A0A096P6D3 A0A2R8ZYN2 A0A2I3SKE0 A0A2K5I5V3 A0A2I2ZRS8 A0A2K5VWS6 A0A1D5QHM2
E2C639 A0A195CS67 A0A067R743 E9JD70 E2ARV1 A0A2J7R0L7 A0A026WAE2 A0A3L8DWH2 Q16KU3 A0A1S4FXA3 W5JAF0 A0A182FFW1 E0VHF1 A0A182TY41 A0A182P3K9 A0A182RFM6 A0A182UVU7 A0A1B0GPC4 A0A182WIQ8 A0A182X6D2 D6WRC2 T1HGC9 Q7QJR1 A0A182KRS0 A0A182I3N8 A0A182QPM5 A0A2K6G9P4 A0A2K6G9P9 A0A182LXR3 A0A182NHM3 A0A2K6G9P1 J3KTB8 A0A1S3GIY4 A0A1D5QY90 L5KPC7 A0A384CCH2 Q3HTU3 A0A2K5CQL4 A0A2R8PBI1 F6R5R2 A0A2K6BQE6 A0A2K5VX29 A0A1B0CRV9 A0A2Y9G5A0 A0A2I3N885 A0A2K6P2V7 A0A2U3YBQ9 A0A182XZ84 G1TYT4 A0A2K6K394 A0A2K5I5V9 A0A2K5XHN5 A0A2I2Y2C2 A0A2R8ZYP4 H2QDM6 F6X3S4 I6L9B3 A0A2K6TDD2 A0A2R8N4Y1 F6QVU8 A0A2K5CQL7 A0A2U3YC18 A0A2K6K3B6 A0A2J8UZC8 A0A2U3YBR9 A0A2K6TDC7 F7F5C6 A0A2U4C3T3 A0A2K5I630 U5L195 P12821-3 A0A2K5CQI8 A0A2K5XHN8 Q9GLN7-2 A0A2R8ZYP1 A0A2U3WBW4 A0A2Y9G4W0 G3RZY8 A0A2K5PZN4 D3DU13 A0A2U3WBV7 A0A2K5VWR1 A0A2K6BQJ6 P12821-4 A0A2K6K3D3 A0A2K6P2X8 A0A096P6D3 A0A2R8ZYN2 A0A2I3SKE0 A0A2K5I5V3 A0A2I2ZRS8 A0A2K5VWS6 A0A1D5QHM2
PDB
6H5W
E-value=7.46976e-29,
Score=312
Ontologies
KEGG
GO
GO:0016020
GO:0008237
GO:0004180
GO:0046872
GO:0008241
GO:0016021
GO:0004129
GO:0005886
GO:0005615
GO:0008238
GO:0071838
GO:0050435
GO:0001822
GO:0005768
GO:0009897
GO:0008240
GO:0060047
GO:0032091
GO:0005764
GO:0042447
GO:0003081
GO:0002446
GO:1903597
GO:0031434
GO:0061098
GO:2000170
GO:0010629
GO:0008144
GO:0070062
GO:0043171
GO:1900086
GO:0031711
GO:1902033
GO:0007283
GO:0050482
GO:0010608
GO:0051019
GO:0060177
GO:0032092
GO:0031404
GO:0008270
GO:0003084
GO:0004175
GO:0008239
GO:0045777
GO:0032943
GO:0003779
GO:0001974
GO:0019229
GO:0006508
GO:0060218
GO:0014910
GO:0002019
GO:0002474
GO:0097746
GO:0005576
GO:0070573
GO:0008217
GO:0002003
GO:0005737
GO:0007178
GO:0004675
GO:0006559
GO:0004887
PANTHER
Topology
Subcellular location
Secreted
Cell membrane Detected in both cell membrane and cytoplasm in neurons. With evidence from 10 publications.
Cytoplasm Detected in both cell membrane and cytoplasm in neurons. With evidence from 10 publications.
Cell membrane Detected in both cell membrane and cytoplasm in neurons. With evidence from 10 publications.
Cytoplasm Detected in both cell membrane and cytoplasm in neurons. With evidence from 10 publications.
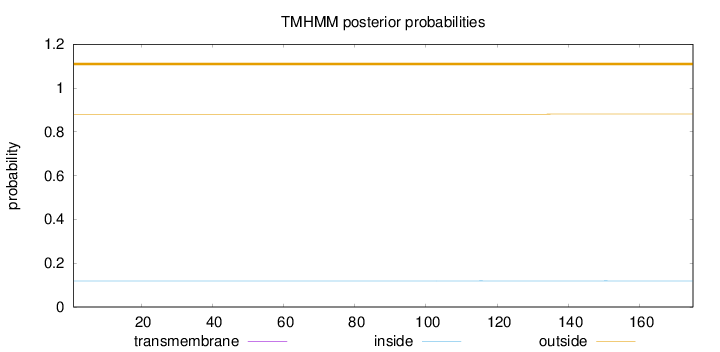
Length:
175
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00247
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11916
outside
1 - 175
Population Genetic Test Statistics
Pi
248.807034
Theta
125.535225
Tajima's D
2.954712
CLR
0.005054
CSRT
0.977601119944003
Interpretation
Uncertain
