Gene
KWMTBOMO00807
Pre Gene Modal
BGIBMGA009737
Annotation
PREDICTED:_coagulation_factor_VII_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.36
Sequence
CDS
ATGGTCCGTTGCAAATCGGAAACTTCAAACAACCTTTTCAATTCCAGATGTGGCGGTTCTTTGATTTCACGTCGTCACGTCGTGACGGCTGGTCACTGTGTGGCCAGAGCCCAGCCCCGCCACGTGAGGGTCACGCTTGGAGACTACGTCATAAACTCGGCGTCGGAACCCTTCCCCGCCTACACGTTCGGAGTGCGGGCCATAAAAGTGCACCCGCTCTTCAAGTTCACACCGCAAGCCGACAGATTCGACGTGGCGGTTCTGACCCTGGACAGAAACGTGCAGTACATGCCCCATATATCCCCGATCTGCCTTCCGGAACGAGGATCAGATTTCCTCGGCCAGTACGGCTGGGCAGCAGGCTGGGGTGCCCTCAGTCCTGGCTCCAGGCTCAGGCCACGGACGCTCCAGGCCGTTGATGTACCGGTTTTGGACAATAGAGTCTGCGAGAGATGGCACAGAGCTAATGGTATAAACGTAGTGATATACCCGGAGATGCTCTGCGCCGGTTACCGGGGGGGAGGCAAGGACAGCTGTCAGGGGGACAGCGGGGGGCCGCTGATGCTGGAACGCGGGGGACGCTGGTACCTCATCGGGGTTGTGTCCGCGGGTTACTCGTGCGCCAGTCGGGGTCAGCCGGGGATCTACCATCGGGTCGCACATACCGTCGACTGGATCTCACACGCCACCACCTTATCATAG
Protein
MVRCKSETSNNLFNSRCGGSLISRRHVVTAGHCVARAQPRHVRVTLGDYVINSASEPFPAYTFGVRAIKVHPLFKFTPQADRFDVAVLTLDRNVQYMPHISPICLPERGSDFLGQYGWAAGWGALSPGSRLRPRTLQAVDVPVLDNRVCERWHRANGINVVIYPEMLCAGYRGGGKDSCQGDSGGPLMLERGGRWYLIGVVSAGYSCASRGQPGIYHRVAHTVDWISHATTLS
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JJN6
A0A2H1V8V1
A0A212FC22
A0A194PZF6
A0A2A4J1X4
A0A194QZJ5
+ More
A0A182TZ67 A0A182MPP3 A0A182WGE3 A0A084W3T7 A0A182I7K8 W5JQE7 A0A182F5Z3 A0A182LMH3 A0A182RHD1 A0A182NBL8 Q16EL5 B0WTZ9 A0A0A9XA11 A0A1S4H0L6 A0A182V8U8 A0A182Y0J6 A0A182QH82 A0A1V1FIT3 A0A182IWC2 A0A336MKZ8 A0A068FB93 A0A2J7RRM7 A0A023F5D8 A0A2S2PLU0 A0A0K8V5L1 A0A1I8MGI8 B4J5E7 A0A1I8NN92 A0A0Q9WA90 A0A067RJY1 A0A0M5J2A3 B4KLP7 W8AQS9 A0A1B0BNH1 A0A1A9XDH2 A0A1B0G148 A0A3Q0JEP0 A0A1I8NN91 B4LLI5 W8AF38 B4N635 A0A0P8Y250 A0A3B0J4M6 B4P3V4 A0A088AM90 A0A0J9TWV4 B3N825 A0A1W4UYC5 Q7JZK6 A0A151IA12 N6W5W3 B3MI89 A0A3B0JP68 A0A0C9R7P4 E2C0S1 A0A1W4UY14 A0A0J9R8I1 B4HSD6 B4QGQ2 A0A2H8TJN1 A1Z7M7 Q28ZD1 B4GHX6 A0A0M9AA41 K7IVN4 A0A310SDW3 A0A3L8DUS7 J9K5X7 A0A2S2QQQ4 A0A1A9UK22 A0A0L7RCK1 E2ADC6 A0A195BFK3 A0A158NRD7 A0A151J5W9 A0A151X5T6 A0A195EXL5 F4WSI9 A0A154P1Q8 A0A182S6D9 N6TN47 A0A1W4X476 A0A139WEY6 A0A1W4XEZ0 E0W195 D6WT60 A0A232ET21 A0A226DEW4 A0A3Q0JID7 A0A1D2MG72 A0A182G5K4 T1HAB6 E9GFB8 A0A0P5M531
A0A182TZ67 A0A182MPP3 A0A182WGE3 A0A084W3T7 A0A182I7K8 W5JQE7 A0A182F5Z3 A0A182LMH3 A0A182RHD1 A0A182NBL8 Q16EL5 B0WTZ9 A0A0A9XA11 A0A1S4H0L6 A0A182V8U8 A0A182Y0J6 A0A182QH82 A0A1V1FIT3 A0A182IWC2 A0A336MKZ8 A0A068FB93 A0A2J7RRM7 A0A023F5D8 A0A2S2PLU0 A0A0K8V5L1 A0A1I8MGI8 B4J5E7 A0A1I8NN92 A0A0Q9WA90 A0A067RJY1 A0A0M5J2A3 B4KLP7 W8AQS9 A0A1B0BNH1 A0A1A9XDH2 A0A1B0G148 A0A3Q0JEP0 A0A1I8NN91 B4LLI5 W8AF38 B4N635 A0A0P8Y250 A0A3B0J4M6 B4P3V4 A0A088AM90 A0A0J9TWV4 B3N825 A0A1W4UYC5 Q7JZK6 A0A151IA12 N6W5W3 B3MI89 A0A3B0JP68 A0A0C9R7P4 E2C0S1 A0A1W4UY14 A0A0J9R8I1 B4HSD6 B4QGQ2 A0A2H8TJN1 A1Z7M7 Q28ZD1 B4GHX6 A0A0M9AA41 K7IVN4 A0A310SDW3 A0A3L8DUS7 J9K5X7 A0A2S2QQQ4 A0A1A9UK22 A0A0L7RCK1 E2ADC6 A0A195BFK3 A0A158NRD7 A0A151J5W9 A0A151X5T6 A0A195EXL5 F4WSI9 A0A154P1Q8 A0A182S6D9 N6TN47 A0A1W4X476 A0A139WEY6 A0A1W4XEZ0 E0W195 D6WT60 A0A232ET21 A0A226DEW4 A0A3Q0JID7 A0A1D2MG72 A0A182G5K4 T1HAB6 E9GFB8 A0A0P5M531
Pubmed
19121390
22118469
26354079
24438588
20920257
23761445
+ More
20966253 17510324 25401762 12364791 25244985 28410430 24952583 25474469 25315136 17994087 18057021 24845553 24495485 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20798317 20075255 30249741 21347285 21719571 23537049 18362917 19820115 20566863 28648823 27289101 26483478 21292972
20966253 17510324 25401762 12364791 25244985 28410430 24952583 25474469 25315136 17994087 18057021 24845553 24495485 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20798317 20075255 30249741 21347285 21719571 23537049 18362917 19820115 20566863 28648823 27289101 26483478 21292972
EMBL
BABH01017455
BABH01017456
ODYU01001260
SOQ37226.1
AGBW02009238
OWR51281.1
+ More
KQ459593 KPI96540.1 NWSH01003593 PCG66075.1 KQ460890 KPJ10963.1 AXCM01011891 ATLV01020134 ATLV01020135 KE525295 KFB44881.1 APCN01003416 ADMH02000814 ETN64964.1 CH478663 EAT32672.1 DS232097 EDS34712.1 GBHO01026052 JAG17552.1 AAAB01008984 AXCN02000678 FX985308 BAX07321.1 UFQT01001434 SSX30545.1 KJ512106 AID60329.1 NEVH01000602 PNF43472.1 GBBI01002368 JAC16344.1 GGMR01017629 MBY30248.1 GDHF01029935 GDHF01018110 JAI22379.1 JAI34204.1 CH916367 EDW01789.1 CH940648 KRF79142.1 KRF79143.1 KK852427 KDR24126.1 CP012524 ALC40781.1 CH933808 EDW08683.2 GAMC01019701 GAMC01019698 GAMC01019697 GAMC01019696 JAB86858.1 JXJN01017387 CCAG010010027 CCAG010010028 EDW59890.1 GAMC01019700 GAMC01019699 JAB86855.1 CH964154 EDW79824.2 CH902619 KPU75895.1 OUUW01000001 SPP74452.1 CM000157 EDW89437.2 KRJ98437.1 KRJ98438.1 CM002911 KMY92403.1 KMY92404.1 KMY92405.1 CH954177 EDV59438.2 AY071201 AE013599 AAL48823.1 ABI31084.1 KQ978249 KYM96006.1 CM000071 ENO01653.1 EDV35934.1 SPP74451.1 GBYB01012384 JAG82151.1 GL451846 EFN78465.1 KMY92402.1 CH480816 EDW47031.1 CM000362 EDX06277.1 GFXV01001683 MBW13488.1 AAF59003.2 EAL25681.2 CH479183 EDW36096.1 KQ435711 KOX79596.1 KQ762882 OAD55357.1 QOIP01000004 RLU24131.1 ABLF02026476 ABLF02026483 GGMS01010864 MBY80067.1 KQ414615 KOC68530.1 GL438749 EFN68542.1 KQ976504 KYM82962.1 ADTU01023935 KQ979928 KYN18609.1 KQ982494 KYQ55746.1 KQ981923 KYN32968.1 GL888322 EGI62854.1 KQ434796 KZC05773.1 APGK01058280 APGK01058281 APGK01058282 APGK01058283 KB741288 ENN70645.1 KQ971354 KYB26530.1 DS235867 EEB19401.1 EFA07412.1 NNAY01002349 OXU21492.1 LNIX01000021 OXA43733.1 LJIJ01001363 ODM91975.1 JXUM01007261 JXUM01007262 KQ560236 KXJ83683.1 ACPB03019345 GL732542 EFX81611.1 GDIQ01164320 JAK87405.1
KQ459593 KPI96540.1 NWSH01003593 PCG66075.1 KQ460890 KPJ10963.1 AXCM01011891 ATLV01020134 ATLV01020135 KE525295 KFB44881.1 APCN01003416 ADMH02000814 ETN64964.1 CH478663 EAT32672.1 DS232097 EDS34712.1 GBHO01026052 JAG17552.1 AAAB01008984 AXCN02000678 FX985308 BAX07321.1 UFQT01001434 SSX30545.1 KJ512106 AID60329.1 NEVH01000602 PNF43472.1 GBBI01002368 JAC16344.1 GGMR01017629 MBY30248.1 GDHF01029935 GDHF01018110 JAI22379.1 JAI34204.1 CH916367 EDW01789.1 CH940648 KRF79142.1 KRF79143.1 KK852427 KDR24126.1 CP012524 ALC40781.1 CH933808 EDW08683.2 GAMC01019701 GAMC01019698 GAMC01019697 GAMC01019696 JAB86858.1 JXJN01017387 CCAG010010027 CCAG010010028 EDW59890.1 GAMC01019700 GAMC01019699 JAB86855.1 CH964154 EDW79824.2 CH902619 KPU75895.1 OUUW01000001 SPP74452.1 CM000157 EDW89437.2 KRJ98437.1 KRJ98438.1 CM002911 KMY92403.1 KMY92404.1 KMY92405.1 CH954177 EDV59438.2 AY071201 AE013599 AAL48823.1 ABI31084.1 KQ978249 KYM96006.1 CM000071 ENO01653.1 EDV35934.1 SPP74451.1 GBYB01012384 JAG82151.1 GL451846 EFN78465.1 KMY92402.1 CH480816 EDW47031.1 CM000362 EDX06277.1 GFXV01001683 MBW13488.1 AAF59003.2 EAL25681.2 CH479183 EDW36096.1 KQ435711 KOX79596.1 KQ762882 OAD55357.1 QOIP01000004 RLU24131.1 ABLF02026476 ABLF02026483 GGMS01010864 MBY80067.1 KQ414615 KOC68530.1 GL438749 EFN68542.1 KQ976504 KYM82962.1 ADTU01023935 KQ979928 KYN18609.1 KQ982494 KYQ55746.1 KQ981923 KYN32968.1 GL888322 EGI62854.1 KQ434796 KZC05773.1 APGK01058280 APGK01058281 APGK01058282 APGK01058283 KB741288 ENN70645.1 KQ971354 KYB26530.1 DS235867 EEB19401.1 EFA07412.1 NNAY01002349 OXU21492.1 LNIX01000021 OXA43733.1 LJIJ01001363 ODM91975.1 JXUM01007261 JXUM01007262 KQ560236 KXJ83683.1 ACPB03019345 GL732542 EFX81611.1 GDIQ01164320 JAK87405.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000053240
UP000075902
+ More
UP000075883 UP000075920 UP000030765 UP000075840 UP000000673 UP000069272 UP000075882 UP000075900 UP000075884 UP000008820 UP000002320 UP000075903 UP000076408 UP000075886 UP000075880 UP000235965 UP000095301 UP000001070 UP000095300 UP000008792 UP000027135 UP000092553 UP000009192 UP000092460 UP000092443 UP000092444 UP000079169 UP000007798 UP000007801 UP000268350 UP000002282 UP000005203 UP000008711 UP000192221 UP000000803 UP000078542 UP000001819 UP000008237 UP000001292 UP000000304 UP000008744 UP000053105 UP000002358 UP000279307 UP000007819 UP000078200 UP000053825 UP000000311 UP000078540 UP000005205 UP000078492 UP000075809 UP000078541 UP000007755 UP000076502 UP000075901 UP000019118 UP000192223 UP000007266 UP000009046 UP000215335 UP000198287 UP000094527 UP000069940 UP000249989 UP000015103 UP000000305
UP000075883 UP000075920 UP000030765 UP000075840 UP000000673 UP000069272 UP000075882 UP000075900 UP000075884 UP000008820 UP000002320 UP000075903 UP000076408 UP000075886 UP000075880 UP000235965 UP000095301 UP000001070 UP000095300 UP000008792 UP000027135 UP000092553 UP000009192 UP000092460 UP000092443 UP000092444 UP000079169 UP000007798 UP000007801 UP000268350 UP000002282 UP000005203 UP000008711 UP000192221 UP000000803 UP000078542 UP000001819 UP000008237 UP000001292 UP000000304 UP000008744 UP000053105 UP000002358 UP000279307 UP000007819 UP000078200 UP000053825 UP000000311 UP000078540 UP000005205 UP000078492 UP000075809 UP000078541 UP000007755 UP000076502 UP000075901 UP000019118 UP000192223 UP000007266 UP000009046 UP000215335 UP000198287 UP000094527 UP000069940 UP000249989 UP000015103 UP000000305
Interpro
Gene 3D
ProteinModelPortal
H9JJN6
A0A2H1V8V1
A0A212FC22
A0A194PZF6
A0A2A4J1X4
A0A194QZJ5
+ More
A0A182TZ67 A0A182MPP3 A0A182WGE3 A0A084W3T7 A0A182I7K8 W5JQE7 A0A182F5Z3 A0A182LMH3 A0A182RHD1 A0A182NBL8 Q16EL5 B0WTZ9 A0A0A9XA11 A0A1S4H0L6 A0A182V8U8 A0A182Y0J6 A0A182QH82 A0A1V1FIT3 A0A182IWC2 A0A336MKZ8 A0A068FB93 A0A2J7RRM7 A0A023F5D8 A0A2S2PLU0 A0A0K8V5L1 A0A1I8MGI8 B4J5E7 A0A1I8NN92 A0A0Q9WA90 A0A067RJY1 A0A0M5J2A3 B4KLP7 W8AQS9 A0A1B0BNH1 A0A1A9XDH2 A0A1B0G148 A0A3Q0JEP0 A0A1I8NN91 B4LLI5 W8AF38 B4N635 A0A0P8Y250 A0A3B0J4M6 B4P3V4 A0A088AM90 A0A0J9TWV4 B3N825 A0A1W4UYC5 Q7JZK6 A0A151IA12 N6W5W3 B3MI89 A0A3B0JP68 A0A0C9R7P4 E2C0S1 A0A1W4UY14 A0A0J9R8I1 B4HSD6 B4QGQ2 A0A2H8TJN1 A1Z7M7 Q28ZD1 B4GHX6 A0A0M9AA41 K7IVN4 A0A310SDW3 A0A3L8DUS7 J9K5X7 A0A2S2QQQ4 A0A1A9UK22 A0A0L7RCK1 E2ADC6 A0A195BFK3 A0A158NRD7 A0A151J5W9 A0A151X5T6 A0A195EXL5 F4WSI9 A0A154P1Q8 A0A182S6D9 N6TN47 A0A1W4X476 A0A139WEY6 A0A1W4XEZ0 E0W195 D6WT60 A0A232ET21 A0A226DEW4 A0A3Q0JID7 A0A1D2MG72 A0A182G5K4 T1HAB6 E9GFB8 A0A0P5M531
A0A182TZ67 A0A182MPP3 A0A182WGE3 A0A084W3T7 A0A182I7K8 W5JQE7 A0A182F5Z3 A0A182LMH3 A0A182RHD1 A0A182NBL8 Q16EL5 B0WTZ9 A0A0A9XA11 A0A1S4H0L6 A0A182V8U8 A0A182Y0J6 A0A182QH82 A0A1V1FIT3 A0A182IWC2 A0A336MKZ8 A0A068FB93 A0A2J7RRM7 A0A023F5D8 A0A2S2PLU0 A0A0K8V5L1 A0A1I8MGI8 B4J5E7 A0A1I8NN92 A0A0Q9WA90 A0A067RJY1 A0A0M5J2A3 B4KLP7 W8AQS9 A0A1B0BNH1 A0A1A9XDH2 A0A1B0G148 A0A3Q0JEP0 A0A1I8NN91 B4LLI5 W8AF38 B4N635 A0A0P8Y250 A0A3B0J4M6 B4P3V4 A0A088AM90 A0A0J9TWV4 B3N825 A0A1W4UYC5 Q7JZK6 A0A151IA12 N6W5W3 B3MI89 A0A3B0JP68 A0A0C9R7P4 E2C0S1 A0A1W4UY14 A0A0J9R8I1 B4HSD6 B4QGQ2 A0A2H8TJN1 A1Z7M7 Q28ZD1 B4GHX6 A0A0M9AA41 K7IVN4 A0A310SDW3 A0A3L8DUS7 J9K5X7 A0A2S2QQQ4 A0A1A9UK22 A0A0L7RCK1 E2ADC6 A0A195BFK3 A0A158NRD7 A0A151J5W9 A0A151X5T6 A0A195EXL5 F4WSI9 A0A154P1Q8 A0A182S6D9 N6TN47 A0A1W4X476 A0A139WEY6 A0A1W4XEZ0 E0W195 D6WT60 A0A232ET21 A0A226DEW4 A0A3Q0JID7 A0A1D2MG72 A0A182G5K4 T1HAB6 E9GFB8 A0A0P5M531
PDB
2F83
E-value=2.44666e-35,
Score=370
Ontologies
GO
Topology
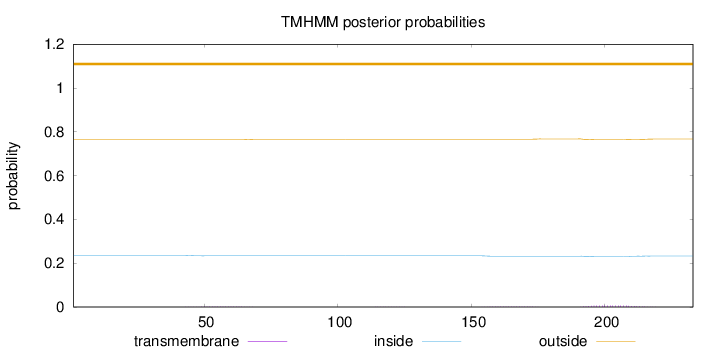
Length:
233
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18548
Exp number, first 60 AAs:
0.02191
Total prob of N-in:
0.23514
outside
1 - 233
Population Genetic Test Statistics
Pi
334.885058
Theta
166.362634
Tajima's D
3.543851
CLR
0.190552
CSRT
0.994650267486626
Interpretation
Uncertain
