Gene
KWMTBOMO00804
Pre Gene Modal
BGIBMGA009694
Annotation
PREDICTED:_serine_proteinase_stubble_isoform_X2_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.62 Nuclear Reliability : 1.266
Sequence
CDS
ATGGACTGTGGTCTACCTTCATCAAGGATACTACAGAAGAGGATCATAGGAGGACGCGAAGCGAGGTTTGCCGAGTTCCCGTGGCAGGCTCACGTCAGGATATCCGAATTTCAATGTGGAGGAGTTTTGGTGTCCCGTTGGTACGTGGCGACGGCGGCTCACTGCGTATCGAGAGCGCGGCCGCGTGACATCGTTGTATGGCTCGGAGCACTCGACACCACGGCTGGCGACGAGACTGCACGGAAACTTGGGGTCGTTCGGAAGATCCTCCATCCTCTGTTTCAATTCCGGATAACCCAACCGGACCGCTACGACATAGCTTTGCTAAAGCTCTCCCAGCCCGTCACGTACATGTCGCATATCCTGCCCATTTGCCTGCCTGATCACGACCTGCAACTGAAAGGCAAGTCGGGAGTTATCGCAGGATGGGGAAAAACGGACACGAGTAATGGCCACACCGGAACGAATCTGTTAAGATCTGCCACCGTGCCTATACTGAGCAAAGAGCAGTGCATCAGCTGGCACCAGAGCAAGCAGATATCAGTCGAGATACATTCAGAGATGATATGCGCCGGGCATTCTGATGGACACCAAGACGCATGTTTAGGTAATAAATGTGAAGTCGGTTTTTTATGGCCACGAAGGCATTAG
Protein
MDCGLPSSRILQKRIIGGREARFAEFPWQAHVRISEFQCGGVLVSRWYVATAAHCVSRARPRDIVVWLGALDTTAGDETARKLGVVRKILHPLFQFRITQPDRYDIALLKLSQPVTYMSHILPICLPDHDLQLKGKSGVIAGWGKTDTSNGHTGTNLLRSATVPILSKEQCISWHQSKQISVEIHSEMICAGHSDGHQDACLGNKCEVGFLWPRRH
Summary
Similarity
Belongs to the peptidase S1 family.
Belongs to the glycosyl hydrolase 18 family.
Belongs to the glycosyl hydrolase 18 family.
Uniprot
H9JJJ3
A0A194PSY2
A0A212EZH1
A0A2A4JBX8
A0A194R133
Q17PV5
+ More
A0A182G5K3 A0A0P6K0Q1 Q1DGX4 A0A1S4EV87 A0A182H5Y5 B0WTZ8 A0A2J7RRK8 W5JNT2 A0A182SLM8 A0A182F5Z2 A0A182UB91 A0A182KG06 A0A182WXP3 A0A182J2A0 A0A084W3T4 A0A182PZH9 A0A336L477 Q7PWE2 A0A182UUN1 A0A182WGE2 A0A034VUP7 A0A1S4H0K2 A0A182NBL7 A0A182RHD0 A0A182LMH1 A0A336L4J3 A0A336MMR9 A0A182Y0J5 A0A182I7K7 A0A336LCS5 A0A182NZG9 A0A0A1WW10 A0A0K8UFY7 A0A1I8MXM3 W8BWD2 B4P3G2 B3N827 A1Z7M5 B4HSD4 A0A1I8Q801 B4QGQ0 A0A1W4UX79 B4LLI7 A0A1B6LKQ3 A0A0M3QUJ4 B4KLP5 B3MI91 A0A3B0J0I1 B4J5E5 B4GHX4 Q28ZD3 B4N633 A0A1B0BNH6 A0A068F5A9 A0A0A9XGC6 A0A1W4XD62 T1HFU7 A0A0A9WT28 D6WT56 A0A0K8SJD7 A0A224XIH6 A0A1J1ICJ4 A0A1A9WYU3 A0A1A9UK22 A0A1B0AFW5 A0A1B0G148 A0A0N8C1U0 A0A0P5YEU9 A0A0P4YRH4 U4TYD6 A0A0P6GPQ1 A0A0P5PCJ2 N6SZG7 A0A0P5JEP8 A0A0P5Z925 E9GRK6 A0A1D2MG43 A0A0P6EY34 A0A0P5QGE0 A0A0N8AE75 A0A0P6EMJ3 A0A0P5LED1 A0A0P5FMR0 A0A0P6FX58 A0A0P5JJY9 A0A0P6F0P6 A0A0P5PN99 A0A0P5Z8G8 A0A0P6DKS9 A0A0P5LCT8 A0A0P6JWL1 A0A0P5UBU8 A0A0P5IKV3 A0A0P6GJ95
A0A182G5K3 A0A0P6K0Q1 Q1DGX4 A0A1S4EV87 A0A182H5Y5 B0WTZ8 A0A2J7RRK8 W5JNT2 A0A182SLM8 A0A182F5Z2 A0A182UB91 A0A182KG06 A0A182WXP3 A0A182J2A0 A0A084W3T4 A0A182PZH9 A0A336L477 Q7PWE2 A0A182UUN1 A0A182WGE2 A0A034VUP7 A0A1S4H0K2 A0A182NBL7 A0A182RHD0 A0A182LMH1 A0A336L4J3 A0A336MMR9 A0A182Y0J5 A0A182I7K7 A0A336LCS5 A0A182NZG9 A0A0A1WW10 A0A0K8UFY7 A0A1I8MXM3 W8BWD2 B4P3G2 B3N827 A1Z7M5 B4HSD4 A0A1I8Q801 B4QGQ0 A0A1W4UX79 B4LLI7 A0A1B6LKQ3 A0A0M3QUJ4 B4KLP5 B3MI91 A0A3B0J0I1 B4J5E5 B4GHX4 Q28ZD3 B4N633 A0A1B0BNH6 A0A068F5A9 A0A0A9XGC6 A0A1W4XD62 T1HFU7 A0A0A9WT28 D6WT56 A0A0K8SJD7 A0A224XIH6 A0A1J1ICJ4 A0A1A9WYU3 A0A1A9UK22 A0A1B0AFW5 A0A1B0G148 A0A0N8C1U0 A0A0P5YEU9 A0A0P4YRH4 U4TYD6 A0A0P6GPQ1 A0A0P5PCJ2 N6SZG7 A0A0P5JEP8 A0A0P5Z925 E9GRK6 A0A1D2MG43 A0A0P6EY34 A0A0P5QGE0 A0A0N8AE75 A0A0P6EMJ3 A0A0P5LED1 A0A0P5FMR0 A0A0P6FX58 A0A0P5JJY9 A0A0P6F0P6 A0A0P5PN99 A0A0P5Z8G8 A0A0P6DKS9 A0A0P5LCT8 A0A0P6JWL1 A0A0P5UBU8 A0A0P5IKV3 A0A0P6GJ95
Pubmed
19121390
26354079
22118469
17510324
26483478
26999592
+ More
20920257 23761445 24438588 12364791 25348373 20966253 25244985 25830018 25315136 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 15632085 24952583 25401762 26823975 18362917 19820115 23537049 21292972 27289101
20920257 23761445 24438588 12364791 25348373 20966253 25244985 25830018 25315136 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 15632085 24952583 25401762 26823975 18362917 19820115 23537049 21292972 27289101
EMBL
BABH01017449
KQ459593
KPI96536.1
AGBW02011281
OWR46899.1
NWSH01002003
+ More
PCG69461.1 KQ460890 KPJ10960.1 CH477189 EAT48736.1 JXUM01007253 KQ560236 KXJ83682.1 GDUN01001001 JAN94918.1 CH900107 EAT32404.2 JXUM01112968 JXUM01112969 KQ565643 KXJ70821.1 DS232097 EDS34711.1 NEVH01000602 PNF43469.1 ADMH02000815 ETN64963.1 ATLV01020131 KE525295 KFB44878.1 AXCN02000678 UFQS01001782 UFQT01001782 SSX12340.1 SSX31791.1 AAAB01008984 EAA14868.4 GAKP01012783 JAC46169.1 SSX12339.1 SSX31790.1 UFQS01001651 UFQT01001651 SSX11735.1 SSX31300.1 APCN01003415 SSX11734.1 SSX31299.1 GBXI01011048 JAD03244.1 GDHF01031874 GDHF01026735 JAI20440.1 JAI25579.1 GAMC01000925 JAC05631.1 CM000157 EDW89435.1 CH954177 EDV59440.1 AE013599 AAF59005.1 CH480816 EDW47029.1 CM000362 CM002911 EDX06275.1 KMY92400.1 CH940648 EDW59892.1 GEBQ01015790 JAT24187.1 CP012524 ALC40779.1 CH933808 EDW08681.1 CH902619 EDV35936.1 OUUW01000001 SPP74454.1 CH916367 EDW01787.1 CH479183 EDW36094.1 CM000071 EAL25680.2 CH964154 EDW79822.1 JXJN01017387 KJ512105 AID60328.1 GBHO01033996 GBHO01033993 GBHO01033990 GBHO01024903 JAG09608.1 JAG09611.1 JAG09614.1 JAG18701.1 ACPB03021400 GBHO01033994 GBHO01033992 GBHO01033991 GBHO01024900 GDHC01021450 JAG09610.1 JAG09612.1 JAG09613.1 JAG18704.1 JAP97178.1 KQ971354 EFA07413.2 GBRD01012570 JAG53254.1 GFTR01005572 JAW10854.1 CVRI01000047 CRK97952.1 CCAG010010027 CCAG010010028 GDIQ01123139 JAL28587.1 GDIP01058856 JAM44859.1 GDIP01224683 JAI98718.1 KB631652 ERL85033.1 GDIQ01032123 JAN62614.1 GDIQ01136432 JAL15294.1 APGK01058276 KB741288 ENN70643.1 GDIQ01225808 JAK25917.1 GDIP01047365 JAM56350.1 GL732560 EFX77790.1 LJIJ01001363 ODM91977.1 GDIQ01068767 JAN25970.1 GDIQ01117492 JAL34234.1 GDIP01156137 JAJ67265.1 GDIQ01073456 JAN21281.1 GDIQ01175256 JAK76469.1 GDIP01146291 JAJ77111.1 GDIQ01041618 JAN53119.1 GDIQ01199005 JAK52720.1 GDIQ01054663 JAN40074.1 GDIQ01146933 JAL04793.1 GDIP01047364 LRGB01002076 JAM56351.1 KZS09397.1 GDIQ01076253 JAN18484.1 GDIQ01171368 JAK80357.1 GDIQ01005680 JAN89057.1 GDIP01116642 JAL87072.1 GDIQ01214256 JAK37469.1 GDIQ01032521 JAN62216.1
PCG69461.1 KQ460890 KPJ10960.1 CH477189 EAT48736.1 JXUM01007253 KQ560236 KXJ83682.1 GDUN01001001 JAN94918.1 CH900107 EAT32404.2 JXUM01112968 JXUM01112969 KQ565643 KXJ70821.1 DS232097 EDS34711.1 NEVH01000602 PNF43469.1 ADMH02000815 ETN64963.1 ATLV01020131 KE525295 KFB44878.1 AXCN02000678 UFQS01001782 UFQT01001782 SSX12340.1 SSX31791.1 AAAB01008984 EAA14868.4 GAKP01012783 JAC46169.1 SSX12339.1 SSX31790.1 UFQS01001651 UFQT01001651 SSX11735.1 SSX31300.1 APCN01003415 SSX11734.1 SSX31299.1 GBXI01011048 JAD03244.1 GDHF01031874 GDHF01026735 JAI20440.1 JAI25579.1 GAMC01000925 JAC05631.1 CM000157 EDW89435.1 CH954177 EDV59440.1 AE013599 AAF59005.1 CH480816 EDW47029.1 CM000362 CM002911 EDX06275.1 KMY92400.1 CH940648 EDW59892.1 GEBQ01015790 JAT24187.1 CP012524 ALC40779.1 CH933808 EDW08681.1 CH902619 EDV35936.1 OUUW01000001 SPP74454.1 CH916367 EDW01787.1 CH479183 EDW36094.1 CM000071 EAL25680.2 CH964154 EDW79822.1 JXJN01017387 KJ512105 AID60328.1 GBHO01033996 GBHO01033993 GBHO01033990 GBHO01024903 JAG09608.1 JAG09611.1 JAG09614.1 JAG18701.1 ACPB03021400 GBHO01033994 GBHO01033992 GBHO01033991 GBHO01024900 GDHC01021450 JAG09610.1 JAG09612.1 JAG09613.1 JAG18704.1 JAP97178.1 KQ971354 EFA07413.2 GBRD01012570 JAG53254.1 GFTR01005572 JAW10854.1 CVRI01000047 CRK97952.1 CCAG010010027 CCAG010010028 GDIQ01123139 JAL28587.1 GDIP01058856 JAM44859.1 GDIP01224683 JAI98718.1 KB631652 ERL85033.1 GDIQ01032123 JAN62614.1 GDIQ01136432 JAL15294.1 APGK01058276 KB741288 ENN70643.1 GDIQ01225808 JAK25917.1 GDIP01047365 JAM56350.1 GL732560 EFX77790.1 LJIJ01001363 ODM91977.1 GDIQ01068767 JAN25970.1 GDIQ01117492 JAL34234.1 GDIP01156137 JAJ67265.1 GDIQ01073456 JAN21281.1 GDIQ01175256 JAK76469.1 GDIP01146291 JAJ77111.1 GDIQ01041618 JAN53119.1 GDIQ01199005 JAK52720.1 GDIQ01054663 JAN40074.1 GDIQ01146933 JAL04793.1 GDIP01047364 LRGB01002076 JAM56351.1 KZS09397.1 GDIQ01076253 JAN18484.1 GDIQ01171368 JAK80357.1 GDIQ01005680 JAN89057.1 GDIP01116642 JAL87072.1 GDIQ01214256 JAK37469.1 GDIQ01032521 JAN62216.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000053240
UP000008820
+ More
UP000069940 UP000249989 UP000002320 UP000235965 UP000000673 UP000075901 UP000069272 UP000075902 UP000075881 UP000076407 UP000075880 UP000030765 UP000075886 UP000007062 UP000075903 UP000075920 UP000075884 UP000075900 UP000075882 UP000076408 UP000075840 UP000075885 UP000095301 UP000002282 UP000008711 UP000000803 UP000001292 UP000095300 UP000000304 UP000192221 UP000008792 UP000092553 UP000009192 UP000007801 UP000268350 UP000001070 UP000008744 UP000001819 UP000007798 UP000092460 UP000192223 UP000015103 UP000007266 UP000183832 UP000091820 UP000078200 UP000092445 UP000092444 UP000030742 UP000019118 UP000000305 UP000094527 UP000076858
UP000069940 UP000249989 UP000002320 UP000235965 UP000000673 UP000075901 UP000069272 UP000075902 UP000075881 UP000076407 UP000075880 UP000030765 UP000075886 UP000007062 UP000075903 UP000075920 UP000075884 UP000075900 UP000075882 UP000076408 UP000075840 UP000075885 UP000095301 UP000002282 UP000008711 UP000000803 UP000001292 UP000095300 UP000000304 UP000192221 UP000008792 UP000092553 UP000009192 UP000007801 UP000268350 UP000001070 UP000008744 UP000001819 UP000007798 UP000092460 UP000192223 UP000015103 UP000007266 UP000183832 UP000091820 UP000078200 UP000092445 UP000092444 UP000030742 UP000019118 UP000000305 UP000094527 UP000076858
Interpro
IPR001314
Peptidase_S1A
+ More
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR033116 TRYPSIN_SER
IPR017853 Glycoside_hydrolase_SF
IPR011583 Chitinase_II
IPR029070 Chitinase_insertion_sf
IPR001223 Glyco_hydro18_cat
IPR018114 TRYPSIN_HIS
IPR013780 Glyco_hydro_b
IPR000322 Glyco_hydro_31
IPR035899 DBL_dom_sf
IPR001849 PH_domain
IPR000219 DH-domain
IPR011993 PH-like_dom_sf
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR033116 TRYPSIN_SER
IPR017853 Glycoside_hydrolase_SF
IPR011583 Chitinase_II
IPR029070 Chitinase_insertion_sf
IPR001223 Glyco_hydro18_cat
IPR018114 TRYPSIN_HIS
IPR013780 Glyco_hydro_b
IPR000322 Glyco_hydro_31
IPR035899 DBL_dom_sf
IPR001849 PH_domain
IPR000219 DH-domain
IPR011993 PH-like_dom_sf
Gene 3D
ProteinModelPortal
H9JJJ3
A0A194PSY2
A0A212EZH1
A0A2A4JBX8
A0A194R133
Q17PV5
+ More
A0A182G5K3 A0A0P6K0Q1 Q1DGX4 A0A1S4EV87 A0A182H5Y5 B0WTZ8 A0A2J7RRK8 W5JNT2 A0A182SLM8 A0A182F5Z2 A0A182UB91 A0A182KG06 A0A182WXP3 A0A182J2A0 A0A084W3T4 A0A182PZH9 A0A336L477 Q7PWE2 A0A182UUN1 A0A182WGE2 A0A034VUP7 A0A1S4H0K2 A0A182NBL7 A0A182RHD0 A0A182LMH1 A0A336L4J3 A0A336MMR9 A0A182Y0J5 A0A182I7K7 A0A336LCS5 A0A182NZG9 A0A0A1WW10 A0A0K8UFY7 A0A1I8MXM3 W8BWD2 B4P3G2 B3N827 A1Z7M5 B4HSD4 A0A1I8Q801 B4QGQ0 A0A1W4UX79 B4LLI7 A0A1B6LKQ3 A0A0M3QUJ4 B4KLP5 B3MI91 A0A3B0J0I1 B4J5E5 B4GHX4 Q28ZD3 B4N633 A0A1B0BNH6 A0A068F5A9 A0A0A9XGC6 A0A1W4XD62 T1HFU7 A0A0A9WT28 D6WT56 A0A0K8SJD7 A0A224XIH6 A0A1J1ICJ4 A0A1A9WYU3 A0A1A9UK22 A0A1B0AFW5 A0A1B0G148 A0A0N8C1U0 A0A0P5YEU9 A0A0P4YRH4 U4TYD6 A0A0P6GPQ1 A0A0P5PCJ2 N6SZG7 A0A0P5JEP8 A0A0P5Z925 E9GRK6 A0A1D2MG43 A0A0P6EY34 A0A0P5QGE0 A0A0N8AE75 A0A0P6EMJ3 A0A0P5LED1 A0A0P5FMR0 A0A0P6FX58 A0A0P5JJY9 A0A0P6F0P6 A0A0P5PN99 A0A0P5Z8G8 A0A0P6DKS9 A0A0P5LCT8 A0A0P6JWL1 A0A0P5UBU8 A0A0P5IKV3 A0A0P6GJ95
A0A182G5K3 A0A0P6K0Q1 Q1DGX4 A0A1S4EV87 A0A182H5Y5 B0WTZ8 A0A2J7RRK8 W5JNT2 A0A182SLM8 A0A182F5Z2 A0A182UB91 A0A182KG06 A0A182WXP3 A0A182J2A0 A0A084W3T4 A0A182PZH9 A0A336L477 Q7PWE2 A0A182UUN1 A0A182WGE2 A0A034VUP7 A0A1S4H0K2 A0A182NBL7 A0A182RHD0 A0A182LMH1 A0A336L4J3 A0A336MMR9 A0A182Y0J5 A0A182I7K7 A0A336LCS5 A0A182NZG9 A0A0A1WW10 A0A0K8UFY7 A0A1I8MXM3 W8BWD2 B4P3G2 B3N827 A1Z7M5 B4HSD4 A0A1I8Q801 B4QGQ0 A0A1W4UX79 B4LLI7 A0A1B6LKQ3 A0A0M3QUJ4 B4KLP5 B3MI91 A0A3B0J0I1 B4J5E5 B4GHX4 Q28ZD3 B4N633 A0A1B0BNH6 A0A068F5A9 A0A0A9XGC6 A0A1W4XD62 T1HFU7 A0A0A9WT28 D6WT56 A0A0K8SJD7 A0A224XIH6 A0A1J1ICJ4 A0A1A9WYU3 A0A1A9UK22 A0A1B0AFW5 A0A1B0G148 A0A0N8C1U0 A0A0P5YEU9 A0A0P4YRH4 U4TYD6 A0A0P6GPQ1 A0A0P5PCJ2 N6SZG7 A0A0P5JEP8 A0A0P5Z925 E9GRK6 A0A1D2MG43 A0A0P6EY34 A0A0P5QGE0 A0A0N8AE75 A0A0P6EMJ3 A0A0P5LED1 A0A0P5FMR0 A0A0P6FX58 A0A0P5JJY9 A0A0P6F0P6 A0A0P5PN99 A0A0P5Z8G8 A0A0P6DKS9 A0A0P5LCT8 A0A0P6JWL1 A0A0P5UBU8 A0A0P5IKV3 A0A0P6GJ95
PDB
4KKD
E-value=1.71234e-23,
Score=267
Ontologies
GO
Topology
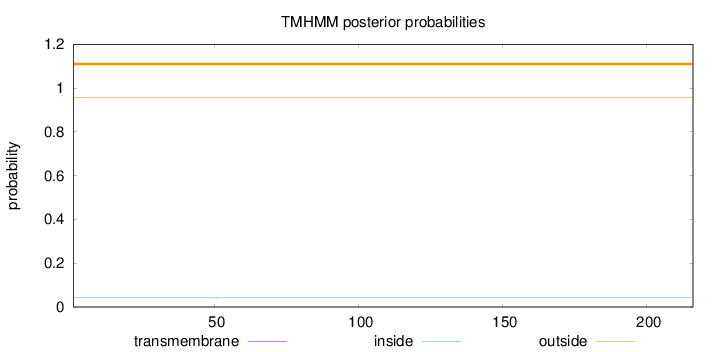
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00918999999999999
Exp number, first 60 AAs:
0.00719
Total prob of N-in:
0.04179
outside
1 - 216
Population Genetic Test Statistics
Pi
66.288442
Theta
51.855324
Tajima's D
-0.679894
CLR
109.510631
CSRT
0.19894005299735
Interpretation
Uncertain
