Gene
KWMTBOMO00794 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009698
Annotation
nascent_polypeptide_associated_complex_protein_alpha_subunit_[Bombyx_mori]
Full name
Nascent polypeptide-associated complex subunit alpha
Alternative Name
Alpha-NAC
Location in the cell
Nuclear Reliability : 2.781
Sequence
CDS
ATGCCAGAACTGACTGAACTTGACAAGGCCACCGCCTCTATGACCGAGAAGCGCAAAGAAGAGACTGCATCATCTGACAGTGACTCTGATGACAACATTCCAGAATTGGAAGATGCAGGTGCACCAGATGCAGGTGGAATCACAAACCCGATTGCTGGAATTGACATCGTCAGCAAATCGAAACAGTCTCGTGGTGAGAAGAAGGCCAGAAAGATAATGAGTAAACTCGGTCTCAAACCAGTACAAGGAGTAGAGAGAGTGACAATCAGAAAATCAAAGAACATTCTCTTTGTCATTAACTCACCTGATGTATACAAGAATCCTCATTCAGACACCTACATTGTTTTTGGTGAAGCCAAGATTGAAGATTTGTCCACACAGGCCACCATGGCTGCAGCAGAGAGATTCAAGGCACCAGAGACCACAGCCACTGGCAATGACGCTTCCACAACTGGAACGACGGTAGCACCAATAGCAGAAGAAGAAGACGAAGAAGGAGTAGACGAGACTGGAGTTGATGAGAAGGACGTGGAGATAGTAATGTCCCAGGCGAACGTCTCTCGAGCGAAAGCAGTCCGAGCGCTCAAGAACAACCAATCTGACATTGTTAATGCAATTATGGAGCTAACAATGTAG
Protein
MPELTELDKATASMTEKRKEETASSDSDSDDNIPELEDAGAPDAGGITNPIAGIDIVSKSKQSRGEKKARKIMSKLGLKPVQGVERVTIRKSKNILFVINSPDVYKNPHSDTYIVFGEAKIEDLSTQATMAAAERFKAPETTATGNDASTTGTTVAPIAEEEDEEGVDETGVDEKDVEIVMSQANVSRAKAVRALKNNQSDIVNAIMELTM
Summary
Description
May promote appropriate targeting of ribosome-nascent polypeptide complexes (By similarity). Required for correct localization of the osk/oskar protein to the posterior pole during embryonic development. The osk protein directs the recruitment of molecules responsible for posterior body patterning and germline formation in the embryo.
Subunit
Part of the nascent polypeptide-associated complex (NAC), consisting of Nac-alpha and bicaudal (bic).
Similarity
Belongs to the NAC-alpha family.
Keywords
Complete proteome
Developmental protein
Protein transport
Reference proteome
Transport
Feature
chain Nascent polypeptide-associated complex subunit alpha
Uniprot
Q1HQ81
H9JJJ7
I4DM86
S4PXV7
I4DJC7
A0A3G1T1N9
+ More
A0A194PU75 A0A194QZI5 A0A212EPN1 A0A1E1W5T5 A0A2H1W3F3 A0A2A4JKN3 A0A2A4ITE7 A0A0K8SSP9 U5EY60 A0A2J7PIS3 A0A1B0CFB5 A0A1L8DIG5 A0A2P8Z4P5 A0A067R157 A0A182N7J5 A0A182V6U9 A0A182L439 A0A182X236 Q7QIB6 A0A182I697 A0A182TN59 A0A182Q980 A0A182PEY4 A0A182WI71 W5JMC0 D6WPS6 A0A1B0DC96 A0A182SGC8 A0A182MJC5 A0A182R9A2 A0A182JDB5 A0A1B6CX80 A0A1B6F758 A0A0K8TP91 A0A1B6EMY6 A0A1B6JWY8 A0A182YJI8 T1DPL2 A0A2M4BZE0 A0A182FI20 A0A1B6I2A0 A0A2M4CND2 A0A2M4A1H2 A0A2M3YZE8 A0A2M3YZH0 E1ZW25 A0A1B6MNW2 V5GCS8 A0A1B6KZF3 E0VJX8 A0A1Q3F7P7 B0WPI0 B4N5Z7 E2BHN8 A0A084WLB0 J3JZJ4 Q16PA5 T1E2Y6 A0A023EJZ8 A0A0J7KZL2 A0A2A3ENP9 V9IJ73 A0A088AUC4 K7IP87 A0A1W4X952 A0A1J1J2V8 A0A0P6IM55 A0A154PG98 A0A0M4E422 A0A0T6B796 B3MG13 A0A1I8PKR6 A0A0P8XP67 F4W5K2 A0A1Y1KA77 B4MDU7 Q0IEK4 A0A1L8EEL8 A0A151X349 A0A0L0C5N2 A0A151JQV6 A0A195D413 A0A1B0BXB1 D3TPE3 A0A1A9XPK0 A0A1A9X4R8 B4P4Y8 A0A0A1WFP3 A0A3B0JSI7 T1PB50 A0A1B0AB83 A0A0B4LEY6 Q94518 B4KQ03
A0A194PU75 A0A194QZI5 A0A212EPN1 A0A1E1W5T5 A0A2H1W3F3 A0A2A4JKN3 A0A2A4ITE7 A0A0K8SSP9 U5EY60 A0A2J7PIS3 A0A1B0CFB5 A0A1L8DIG5 A0A2P8Z4P5 A0A067R157 A0A182N7J5 A0A182V6U9 A0A182L439 A0A182X236 Q7QIB6 A0A182I697 A0A182TN59 A0A182Q980 A0A182PEY4 A0A182WI71 W5JMC0 D6WPS6 A0A1B0DC96 A0A182SGC8 A0A182MJC5 A0A182R9A2 A0A182JDB5 A0A1B6CX80 A0A1B6F758 A0A0K8TP91 A0A1B6EMY6 A0A1B6JWY8 A0A182YJI8 T1DPL2 A0A2M4BZE0 A0A182FI20 A0A1B6I2A0 A0A2M4CND2 A0A2M4A1H2 A0A2M3YZE8 A0A2M3YZH0 E1ZW25 A0A1B6MNW2 V5GCS8 A0A1B6KZF3 E0VJX8 A0A1Q3F7P7 B0WPI0 B4N5Z7 E2BHN8 A0A084WLB0 J3JZJ4 Q16PA5 T1E2Y6 A0A023EJZ8 A0A0J7KZL2 A0A2A3ENP9 V9IJ73 A0A088AUC4 K7IP87 A0A1W4X952 A0A1J1J2V8 A0A0P6IM55 A0A154PG98 A0A0M4E422 A0A0T6B796 B3MG13 A0A1I8PKR6 A0A0P8XP67 F4W5K2 A0A1Y1KA77 B4MDU7 Q0IEK4 A0A1L8EEL8 A0A151X349 A0A0L0C5N2 A0A151JQV6 A0A195D413 A0A1B0BXB1 D3TPE3 A0A1A9XPK0 A0A1A9X4R8 B4P4Y8 A0A0A1WFP3 A0A3B0JSI7 T1PB50 A0A1B0AB83 A0A0B4LEY6 Q94518 B4KQ03
Pubmed
19121390
22651552
23622113
26354079
22118469
29403074
+ More
24845553 20966253 12364791 14747013 17210077 20920257 23761445 18362917 19820115 26369729 25244985 20798317 20566863 17994087 24438588 22516182 23537049 17510324 24330624 24945155 26483478 20075255 18057021 21719571 28004739 26108605 20353571 17550304 25830018 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10071211 11102369 12537569 15239960
24845553 20966253 12364791 14747013 17210077 20920257 23761445 18362917 19820115 26369729 25244985 20798317 20566863 17994087 24438588 22516182 23537049 17510324 24330624 24945155 26483478 20075255 18057021 21719571 28004739 26108605 20353571 17550304 25830018 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10071211 11102369 12537569 15239960
EMBL
DQ443171
ABF51260.1
BABH01017422
AK402404
BAM19026.1
GAIX01004368
+ More
JAA88192.1 AK401395 BAM18017.1 MG992444 AXY94882.1 KQ459593 KPI96528.1 KQ460890 KPJ10953.1 AGBW02013449 OWR43417.1 GDQN01008719 JAT82335.1 ODYU01006085 SOQ47620.1 NWSH01001257 PCG71960.1 NWSH01008684 PCG62452.1 GBRD01009625 JAG56199.1 GANO01000507 JAB59364.1 NEVH01024966 PNF16240.1 AJWK01009907 GFDF01007927 JAV06157.1 PYGN01000196 PSN51463.1 KK869083 KDR11265.1 AAAB01008807 EAA04708.2 APCN01003599 AXCN02000576 ADMH02000632 ETN65522.1 KQ971354 EFA06181.1 AJVK01030690 AXCM01006814 GEDC01019270 JAS18028.1 GECZ01023732 GECZ01013407 JAS46037.1 JAS56362.1 GDAI01001439 JAI16164.1 GECZ01030513 JAS39256.1 GECU01004303 JAT03404.1 GAMD01003254 JAA98336.1 GGFJ01009309 MBW58450.1 GECU01026648 JAS81058.1 GGFL01002523 MBW66701.1 GGFK01001249 MBW34570.1 GGFM01000882 MBW21633.1 GGFM01000895 MBW21646.1 GL434758 EFN74612.1 GEBQ01002347 JAT37630.1 GALX01000541 JAB67925.1 GEBQ01023155 JAT16822.1 DS235231 EEB13684.1 GFDL01011478 JAV23567.1 DS232024 EDS32368.1 CH964154 EDW79786.1 GL448322 EFN84786.1 ATLV01024210 KE525350 KFB51004.1 APGK01034946 BT128676 KB740923 KB631963 AEE63633.1 ENN78092.1 ERL87496.1 CH477789 EAT36197.1 GALA01000865 JAA93987.1 JXUM01078212 JXUM01078213 JXUM01078214 GAPW01004178 KQ563051 JAC09420.1 KXJ74566.1 LBMM01001759 KMQ95806.1 KZ288205 PBC33134.1 JR047348 AEY60389.1 CVRI01000067 CRL06703.1 GDIQ01003438 JAN91299.1 KQ434899 KZC10885.1 CP012524 ALC40691.1 LJIG01009364 KRT83228.1 CH902619 EDV36708.1 KPU76374.1 KPU76375.1 GL887655 EGI70499.1 GEZM01087975 JAV58363.1 CH940662 EDW58712.1 CH477600 EAT38459.1 GFDG01001632 JAV17167.1 KQ982562 KYQ54813.1 JRES01000960 KNC26734.1 KQ978642 KYN29500.1 KQ976881 KYN07586.1 JXJN01022150 EZ423295 ADD19571.1 CM000158 EDW90709.1 KRJ99384.1 GBXI01017059 JAC97232.1 OUUW01000001 SPP74058.1 KA645375 AFP60004.1 AE013599 AHN56161.1 Y08969 AF017783 AY075332 CH933808 EDW08105.1
JAA88192.1 AK401395 BAM18017.1 MG992444 AXY94882.1 KQ459593 KPI96528.1 KQ460890 KPJ10953.1 AGBW02013449 OWR43417.1 GDQN01008719 JAT82335.1 ODYU01006085 SOQ47620.1 NWSH01001257 PCG71960.1 NWSH01008684 PCG62452.1 GBRD01009625 JAG56199.1 GANO01000507 JAB59364.1 NEVH01024966 PNF16240.1 AJWK01009907 GFDF01007927 JAV06157.1 PYGN01000196 PSN51463.1 KK869083 KDR11265.1 AAAB01008807 EAA04708.2 APCN01003599 AXCN02000576 ADMH02000632 ETN65522.1 KQ971354 EFA06181.1 AJVK01030690 AXCM01006814 GEDC01019270 JAS18028.1 GECZ01023732 GECZ01013407 JAS46037.1 JAS56362.1 GDAI01001439 JAI16164.1 GECZ01030513 JAS39256.1 GECU01004303 JAT03404.1 GAMD01003254 JAA98336.1 GGFJ01009309 MBW58450.1 GECU01026648 JAS81058.1 GGFL01002523 MBW66701.1 GGFK01001249 MBW34570.1 GGFM01000882 MBW21633.1 GGFM01000895 MBW21646.1 GL434758 EFN74612.1 GEBQ01002347 JAT37630.1 GALX01000541 JAB67925.1 GEBQ01023155 JAT16822.1 DS235231 EEB13684.1 GFDL01011478 JAV23567.1 DS232024 EDS32368.1 CH964154 EDW79786.1 GL448322 EFN84786.1 ATLV01024210 KE525350 KFB51004.1 APGK01034946 BT128676 KB740923 KB631963 AEE63633.1 ENN78092.1 ERL87496.1 CH477789 EAT36197.1 GALA01000865 JAA93987.1 JXUM01078212 JXUM01078213 JXUM01078214 GAPW01004178 KQ563051 JAC09420.1 KXJ74566.1 LBMM01001759 KMQ95806.1 KZ288205 PBC33134.1 JR047348 AEY60389.1 CVRI01000067 CRL06703.1 GDIQ01003438 JAN91299.1 KQ434899 KZC10885.1 CP012524 ALC40691.1 LJIG01009364 KRT83228.1 CH902619 EDV36708.1 KPU76374.1 KPU76375.1 GL887655 EGI70499.1 GEZM01087975 JAV58363.1 CH940662 EDW58712.1 CH477600 EAT38459.1 GFDG01001632 JAV17167.1 KQ982562 KYQ54813.1 JRES01000960 KNC26734.1 KQ978642 KYN29500.1 KQ976881 KYN07586.1 JXJN01022150 EZ423295 ADD19571.1 CM000158 EDW90709.1 KRJ99384.1 GBXI01017059 JAC97232.1 OUUW01000001 SPP74058.1 KA645375 AFP60004.1 AE013599 AHN56161.1 Y08969 AF017783 AY075332 CH933808 EDW08105.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000235965
+ More
UP000092461 UP000245037 UP000027135 UP000075884 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075902 UP000075886 UP000075885 UP000075920 UP000000673 UP000007266 UP000092462 UP000075901 UP000075883 UP000075900 UP000075880 UP000076408 UP000069272 UP000000311 UP000009046 UP000002320 UP000007798 UP000008237 UP000030765 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000036403 UP000242457 UP000005203 UP000002358 UP000192223 UP000183832 UP000076502 UP000092553 UP000007801 UP000095300 UP000007755 UP000008792 UP000075809 UP000037069 UP000078492 UP000078542 UP000092460 UP000092443 UP000091820 UP000002282 UP000268350 UP000095301 UP000092445 UP000000803 UP000009192
UP000092461 UP000245037 UP000027135 UP000075884 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075902 UP000075886 UP000075885 UP000075920 UP000000673 UP000007266 UP000092462 UP000075901 UP000075883 UP000075900 UP000075880 UP000076408 UP000069272 UP000000311 UP000009046 UP000002320 UP000007798 UP000008237 UP000030765 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000036403 UP000242457 UP000005203 UP000002358 UP000192223 UP000183832 UP000076502 UP000092553 UP000007801 UP000095300 UP000007755 UP000008792 UP000075809 UP000037069 UP000078492 UP000078542 UP000092460 UP000092443 UP000091820 UP000002282 UP000268350 UP000095301 UP000092445 UP000000803 UP000009192
Pfam
PF01849 NAC
Gene 3D
ProteinModelPortal
Q1HQ81
H9JJJ7
I4DM86
S4PXV7
I4DJC7
A0A3G1T1N9
+ More
A0A194PU75 A0A194QZI5 A0A212EPN1 A0A1E1W5T5 A0A2H1W3F3 A0A2A4JKN3 A0A2A4ITE7 A0A0K8SSP9 U5EY60 A0A2J7PIS3 A0A1B0CFB5 A0A1L8DIG5 A0A2P8Z4P5 A0A067R157 A0A182N7J5 A0A182V6U9 A0A182L439 A0A182X236 Q7QIB6 A0A182I697 A0A182TN59 A0A182Q980 A0A182PEY4 A0A182WI71 W5JMC0 D6WPS6 A0A1B0DC96 A0A182SGC8 A0A182MJC5 A0A182R9A2 A0A182JDB5 A0A1B6CX80 A0A1B6F758 A0A0K8TP91 A0A1B6EMY6 A0A1B6JWY8 A0A182YJI8 T1DPL2 A0A2M4BZE0 A0A182FI20 A0A1B6I2A0 A0A2M4CND2 A0A2M4A1H2 A0A2M3YZE8 A0A2M3YZH0 E1ZW25 A0A1B6MNW2 V5GCS8 A0A1B6KZF3 E0VJX8 A0A1Q3F7P7 B0WPI0 B4N5Z7 E2BHN8 A0A084WLB0 J3JZJ4 Q16PA5 T1E2Y6 A0A023EJZ8 A0A0J7KZL2 A0A2A3ENP9 V9IJ73 A0A088AUC4 K7IP87 A0A1W4X952 A0A1J1J2V8 A0A0P6IM55 A0A154PG98 A0A0M4E422 A0A0T6B796 B3MG13 A0A1I8PKR6 A0A0P8XP67 F4W5K2 A0A1Y1KA77 B4MDU7 Q0IEK4 A0A1L8EEL8 A0A151X349 A0A0L0C5N2 A0A151JQV6 A0A195D413 A0A1B0BXB1 D3TPE3 A0A1A9XPK0 A0A1A9X4R8 B4P4Y8 A0A0A1WFP3 A0A3B0JSI7 T1PB50 A0A1B0AB83 A0A0B4LEY6 Q94518 B4KQ03
A0A194PU75 A0A194QZI5 A0A212EPN1 A0A1E1W5T5 A0A2H1W3F3 A0A2A4JKN3 A0A2A4ITE7 A0A0K8SSP9 U5EY60 A0A2J7PIS3 A0A1B0CFB5 A0A1L8DIG5 A0A2P8Z4P5 A0A067R157 A0A182N7J5 A0A182V6U9 A0A182L439 A0A182X236 Q7QIB6 A0A182I697 A0A182TN59 A0A182Q980 A0A182PEY4 A0A182WI71 W5JMC0 D6WPS6 A0A1B0DC96 A0A182SGC8 A0A182MJC5 A0A182R9A2 A0A182JDB5 A0A1B6CX80 A0A1B6F758 A0A0K8TP91 A0A1B6EMY6 A0A1B6JWY8 A0A182YJI8 T1DPL2 A0A2M4BZE0 A0A182FI20 A0A1B6I2A0 A0A2M4CND2 A0A2M4A1H2 A0A2M3YZE8 A0A2M3YZH0 E1ZW25 A0A1B6MNW2 V5GCS8 A0A1B6KZF3 E0VJX8 A0A1Q3F7P7 B0WPI0 B4N5Z7 E2BHN8 A0A084WLB0 J3JZJ4 Q16PA5 T1E2Y6 A0A023EJZ8 A0A0J7KZL2 A0A2A3ENP9 V9IJ73 A0A088AUC4 K7IP87 A0A1W4X952 A0A1J1J2V8 A0A0P6IM55 A0A154PG98 A0A0M4E422 A0A0T6B796 B3MG13 A0A1I8PKR6 A0A0P8XP67 F4W5K2 A0A1Y1KA77 B4MDU7 Q0IEK4 A0A1L8EEL8 A0A151X349 A0A0L0C5N2 A0A151JQV6 A0A195D413 A0A1B0BXB1 D3TPE3 A0A1A9XPK0 A0A1A9X4R8 B4P4Y8 A0A0A1WFP3 A0A3B0JSI7 T1PB50 A0A1B0AB83 A0A0B4LEY6 Q94518 B4KQ03
PDB
3MCE
E-value=2.68184e-16,
Score=205
Ontologies
PANTHER
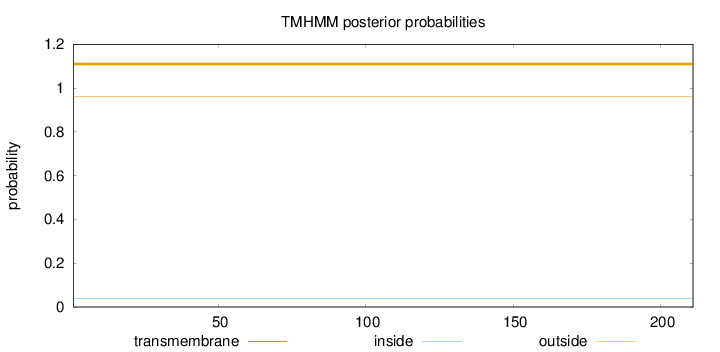
Topology
Length:
211
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00061
Exp number, first 60 AAs:
0.00061
Total prob of N-in:
0.03773
outside
1 - 211
Population Genetic Test Statistics
Pi
329.954209
Theta
203.607086
Tajima's D
1.602194
CLR
0.008404
CSRT
0.815509224538773
Interpretation
Uncertain
