Gene
KWMTBOMO00793 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009699
Annotation
PREDICTED:_T-complex_protein_1_subunit_epsilon_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.754
Sequence
CDS
ATGGCCCTGTTCCCGGGATCGGTGGCCTTTGATGAGTATGGCCGTCCATTTATAATCTTGAGGGATCAAGATAGACAGAAAAGACTTACCGGTACCGATGCCATAAAGTCACATATACAGGCAGCCCGACAAATTGCAAGCACTCTTCGCACATCTCTTGGTCCCCGCGGTCTTGATAAGCTTATGGTGTCGTCTGATGGTGATGTGACAGTAACAAATGATGGGGCTACAATTCTAAAAATGATGGACGTTGAGCATCAGATTGGTAAACTACTTGTGCAATTGGCTCAGAGTCAAGATGACGAAATTGGTGATGGTACCACAGGGGTTGTTGTTCTTGCTGGTGCACTCCTTGAGCAAGCCTCAAATCTGTTGGACAAAGGTATCCATCCGATTCGTATAGCCGATGGTTATGAGATGGCTGCCGCCCTGGCACTGACCCATCTTGACAGCGTCAGTGAAGCTTTCCCAGTCAACAAGAATACGAGGGAAAATTTAATTAAGGTGGCCATGACAACCTTGGGCAGTAAAGTGGTGGTCAAGTGTCACAGGTTGATGGCAGAAATAGCTGTTGATGCTGTCCTGGCTGTGGCAGACTTAGAGACGCGAGACGTCAATTTCGAGCTGATCAAAGTGGAGGGAAAAGTTGGAGGACGCATGGAGGATTCTGTTCTTGTTCGAGGTGTCGTTGTGGACAAAACGATGAGTCACCCTCAAATGCCCAAAGTACTCCGGAATGTGAAACTAGCAATCCTGACCTGCCCGTTCGAACCACCCAAACCCAAGACCAAGCATAAGCTCCAGGTGGGCTCCGCTGATGAATACCGTGAGCTGAGACAGTATGAACACGATAAATTCATCGAAATGGTCCGCAAAGTTAAGGATGCAGGTGCTACTCTAGCAATCTGCCAGTGGGGCTTTGATGATGAAGCCAACCATCTCCTGCTGGCTGAAGGTCTACCAGCCGTACGCTGGGTGGGTGGACCAGAAATGGAGCTCATCGCCATTGCGACTGGCGGCAGGATCGTGCCTAGATTTGAAGAACTCACTCCAGAGAAACTTGGCTCATGTGGACTAGTACGAGAACTTACTTTTGGTACATCAAAAGAAGAGATGTTGGTGATAGAAGAGTGTTCCAACTCTCGAGCGGTAACTGTACTTGTGAGAGGAGGGAATAGAATGATTGTGGAGGAAGCCAAGCGTTCGGTGCATGATGCTCTGTGCATTGTGCGCAGTCTTGTACAGGACTCGCGCGTGGTGTACGGCGGTGGCGCGGCGGAGGCGTCGTGCTCGCTGGCAGTCAGCGCGGCCGCTGAGAAGCTATCCTCGCTCGATCAGTACGCGTTCAGAGCCTTCGGAGATGCGCTCGAGGCCATACCTCTGGCACTCGCCGAAAACAGCGGCTTATCACCTATTGATGCATTGTCTGAAGTGAAAGCGAGACAAGTTGCTGAGAATAATCCCTACTTGGGAATCGACTGCATGTCAAAAGGTTCAAATGACATGAAAGCAATGAACGTAATTGAATCGTTGCACTCAAAGAAACAGCAGATAGCTCTGGCCACCCAACTCGTCAAGATGATACTCAAGATTGATGATGTCCGGTCACCTGCAGACTCCATCGAATAA
Protein
MALFPGSVAFDEYGRPFIILRDQDRQKRLTGTDAIKSHIQAARQIASTLRTSLGPRGLDKLMVSSDGDVTVTNDGATILKMMDVEHQIGKLLVQLAQSQDDEIGDGTTGVVVLAGALLEQASNLLDKGIHPIRIADGYEMAAALALTHLDSVSEAFPVNKNTRENLIKVAMTTLGSKVVVKCHRLMAEIAVDAVLAVADLETRDVNFELIKVEGKVGGRMEDSVLVRGVVVDKTMSHPQMPKVLRNVKLAILTCPFEPPKPKTKHKLQVGSADEYRELRQYEHDKFIEMVRKVKDAGATLAICQWGFDDEANHLLLAEGLPAVRWVGGPEMELIAIATGGRIVPRFEELTPEKLGSCGLVRELTFGTSKEEMLVIEECSNSRAVTVLVRGGNRMIVEEAKRSVHDALCIVRSLVQDSRVVYGGGAAEASCSLAVSAAAEKLSSLDQYAFRAFGDALEAIPLALAENSGLSPIDALSEVKARQVAENNPYLGIDCMSKGSNDMKAMNVIESLHSKKQQIALATQLVKMILKIDDVRSPADSIE
Summary
Similarity
Belongs to the TCP-1 chaperonin family.
Uniprot
H9JJJ8
A0A2A4JJQ5
A0A194PT86
A0A194QZS2
A0A212EPJ9
A0A1B6KJV8
+ More
A0A023EWD6 A0A097PHN6 A0A1B6GHV6 A0A1L8EE66 A0A2P8Y706 A0A182H9M4 A0A1S4EZR6 T1PAU1 A0A0L0CF33 B4KMN2 A0A3B0J741 Q17KD5 Q293E0 B4GBU9 B4J4M7 A0A232FBI2 A0A1I8PBX8 W8C6G9 B0XGB7 K7IV31 T1D3W9 B4HP56 A0A0J9RAR7 B3NS73 Q7KKI0 A0A1W4ULS5 A0A1W4WN70 A0A182Y3I7 B3MH19 A0A2M4A6N5 A0A1A9XDF7 A0A1B0AML6 A0A2J7RAN2 B4LNH7 A0A182NU32 A0A1A9UK11 T1E783 D2A1F8 A0A2M4A6E1 A0A2M4BJH8 A0A182F140 A0A1B0AFU9 B4P5N3 W5J3N6 E0VSM9 A0A1Q3F539 A0A1Q3F4J5 A0A0A1X9V6 A0A0M3QVL0 A0A034W3H8 U5EUG1 A0A1J1IHK9 B4NMW6 A0A182W6S4 A0A182XL29 A0A182UUS1 A0A182U1C1 A0A182PCM4 A0A0K8TSU0 A0A2M4BKI2 A0A1B0D4S6 A0A1L8DUF7 A0A182Q8N6 A0A182J8P6 A0A182JXW2 A0A084W4S8 A0A182L112 Q7Q467 A0A182ICJ3 A0A0U2HUG4 A0A182M0Q1 X1WV13 A0A182R2Y6 A0A0P6H8C7 A0A0N8AI12 A0A0N8CEK0 A0A1Y1MKQ3 A0A2S2N8L1 A0A0P4ZYX3 A0A1B6D5X1 A0A0N7ZQT6 A0A0P5I5M7 A0A067RTG8 A0A0P5H4B5 A0A154P7A5 N6TEJ1 A0A336MAX3 J3JWI9 N6TRW5 E9H224 A0A0P4WBF7 T1IZH6 V5RDW2 A0A0T6B8L4
A0A023EWD6 A0A097PHN6 A0A1B6GHV6 A0A1L8EE66 A0A2P8Y706 A0A182H9M4 A0A1S4EZR6 T1PAU1 A0A0L0CF33 B4KMN2 A0A3B0J741 Q17KD5 Q293E0 B4GBU9 B4J4M7 A0A232FBI2 A0A1I8PBX8 W8C6G9 B0XGB7 K7IV31 T1D3W9 B4HP56 A0A0J9RAR7 B3NS73 Q7KKI0 A0A1W4ULS5 A0A1W4WN70 A0A182Y3I7 B3MH19 A0A2M4A6N5 A0A1A9XDF7 A0A1B0AML6 A0A2J7RAN2 B4LNH7 A0A182NU32 A0A1A9UK11 T1E783 D2A1F8 A0A2M4A6E1 A0A2M4BJH8 A0A182F140 A0A1B0AFU9 B4P5N3 W5J3N6 E0VSM9 A0A1Q3F539 A0A1Q3F4J5 A0A0A1X9V6 A0A0M3QVL0 A0A034W3H8 U5EUG1 A0A1J1IHK9 B4NMW6 A0A182W6S4 A0A182XL29 A0A182UUS1 A0A182U1C1 A0A182PCM4 A0A0K8TSU0 A0A2M4BKI2 A0A1B0D4S6 A0A1L8DUF7 A0A182Q8N6 A0A182J8P6 A0A182JXW2 A0A084W4S8 A0A182L112 Q7Q467 A0A182ICJ3 A0A0U2HUG4 A0A182M0Q1 X1WV13 A0A182R2Y6 A0A0P6H8C7 A0A0N8AI12 A0A0N8CEK0 A0A1Y1MKQ3 A0A2S2N8L1 A0A0P4ZYX3 A0A1B6D5X1 A0A0N7ZQT6 A0A0P5I5M7 A0A067RTG8 A0A0P5H4B5 A0A154P7A5 N6TEJ1 A0A336MAX3 J3JWI9 N6TRW5 E9H224 A0A0P4WBF7 T1IZH6 V5RDW2 A0A0T6B8L4
Pubmed
19121390
26354079
22118469
24945155
29403074
26483478
+ More
25315136 26108605 17994087 17510324 15632085 28648823 24495485 20075255 24330624 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 18362917 19820115 17550304 20920257 23761445 20566863 25830018 25348373 26369729 24438588 20966253 12364791 14747013 17210077 28004739 24845553 23537049 22516182 21292972
25315136 26108605 17994087 17510324 15632085 28648823 24495485 20075255 24330624 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 18362917 19820115 17550304 20920257 23761445 20566863 25830018 25348373 26369729 24438588 20966253 12364791 14747013 17210077 28004739 24845553 23537049 22516182 21292972
EMBL
BABH01017422
NWSH01001257
PCG71958.1
KQ459593
KPI96527.1
KQ460890
+ More
KPJ10952.1 AGBW02013449 OWR43418.1 GEBQ01028251 GEBQ01026767 JAT11726.1 JAT13210.1 GAPW01000864 JAC12734.1 KM221891 AIU47032.1 GECZ01023670 GECZ01022974 GECZ01021022 GECZ01017591 GECZ01007876 GECZ01003617 JAS46099.1 JAS46795.1 JAS48747.1 JAS52178.1 JAS61893.1 JAS66152.1 GFDG01001910 JAV16889.1 PYGN01000852 PSN40030.1 JXUM01030590 KQ560889 KXJ80468.1 KA645857 AFP60486.1 JRES01000487 KNC30851.1 CH933808 EDW08774.1 OUUW01000001 SPP75923.1 CH477225 EAT47156.1 CM000071 EAL24671.1 CH479181 EDW31328.1 CH916367 EDW02732.1 NNAY01000532 OXU27840.1 GAMC01001122 JAC05434.1 DS233011 EDS27370.1 AAZX01001194 GALA01001203 JAA93649.1 CH480816 EDW47505.1 CM002911 KMY93110.1 KMY93111.1 CH954179 EDV56375.1 AF172635 AE013599 AAD46928.1 AAF58565.1 CH902619 EDV35778.1 GGFK01003090 MBW36411.1 JXJN01000499 NEVH01006567 PNF37886.1 CH940648 EDW62157.1 GAMD01003272 JAA98318.1 KQ971338 EFA02100.1 GGFK01003032 MBW36353.1 GGFJ01004085 MBW53226.1 CM000158 EDW90830.1 ADMH02002106 ETN58977.1 DS235751 EEB16385.1 GFDL01012370 JAV22675.1 GFDL01012565 JAV22480.1 GBXI01006208 JAD08084.1 CP012524 ALC42589.1 GAKP01010629 JAC48323.1 GANO01001488 JAB58383.1 CVRI01000047 CRK97937.1 CH964282 EDW85705.1 GDAI01000407 JAI17196.1 GGFJ01004157 MBW53298.1 AJVK01024828 GFDF01004013 JAV10071.1 AXCN02000781 ATLV01020380 KE525299 KFB45222.1 AAAB01008964 EAA12349.3 APCN01000802 KR181953 ALJ76677.1 AXCM01007289 ABLF02022344 ABLF02022348 ABLF02045642 GDIQ01023215 JAN71522.1 GDIP01145441 JAJ77961.1 GDIP01252174 GDIP01230534 GDIP01139447 GDIP01135351 LRGB01001005 JAL64267.1 KZS14107.1 GEZM01028452 JAV86299.1 GGMR01000850 MBY13469.1 GDIP01219287 JAJ04115.1 GEDC01016225 JAS21073.1 GDIP01221649 JAJ01753.1 GDIQ01224399 JAK27326.1 KK852427 KDR24110.1 GDIQ01239244 JAK12481.1 KQ434822 KZC07078.1 APGK01032099 KB740835 ENN78789.1 UFQT01000827 UFQT01002674 SSX27425.1 SSX33909.1 BT127607 AEE62569.1 APGK01005323 KB736187 ENN83244.1 GL732585 EFX74207.1 GDRN01066335 JAI64549.1 AFFK01020457 JH431580 KF621131 AHB33467.1 LJIG01009390 KRT83175.1
KPJ10952.1 AGBW02013449 OWR43418.1 GEBQ01028251 GEBQ01026767 JAT11726.1 JAT13210.1 GAPW01000864 JAC12734.1 KM221891 AIU47032.1 GECZ01023670 GECZ01022974 GECZ01021022 GECZ01017591 GECZ01007876 GECZ01003617 JAS46099.1 JAS46795.1 JAS48747.1 JAS52178.1 JAS61893.1 JAS66152.1 GFDG01001910 JAV16889.1 PYGN01000852 PSN40030.1 JXUM01030590 KQ560889 KXJ80468.1 KA645857 AFP60486.1 JRES01000487 KNC30851.1 CH933808 EDW08774.1 OUUW01000001 SPP75923.1 CH477225 EAT47156.1 CM000071 EAL24671.1 CH479181 EDW31328.1 CH916367 EDW02732.1 NNAY01000532 OXU27840.1 GAMC01001122 JAC05434.1 DS233011 EDS27370.1 AAZX01001194 GALA01001203 JAA93649.1 CH480816 EDW47505.1 CM002911 KMY93110.1 KMY93111.1 CH954179 EDV56375.1 AF172635 AE013599 AAD46928.1 AAF58565.1 CH902619 EDV35778.1 GGFK01003090 MBW36411.1 JXJN01000499 NEVH01006567 PNF37886.1 CH940648 EDW62157.1 GAMD01003272 JAA98318.1 KQ971338 EFA02100.1 GGFK01003032 MBW36353.1 GGFJ01004085 MBW53226.1 CM000158 EDW90830.1 ADMH02002106 ETN58977.1 DS235751 EEB16385.1 GFDL01012370 JAV22675.1 GFDL01012565 JAV22480.1 GBXI01006208 JAD08084.1 CP012524 ALC42589.1 GAKP01010629 JAC48323.1 GANO01001488 JAB58383.1 CVRI01000047 CRK97937.1 CH964282 EDW85705.1 GDAI01000407 JAI17196.1 GGFJ01004157 MBW53298.1 AJVK01024828 GFDF01004013 JAV10071.1 AXCN02000781 ATLV01020380 KE525299 KFB45222.1 AAAB01008964 EAA12349.3 APCN01000802 KR181953 ALJ76677.1 AXCM01007289 ABLF02022344 ABLF02022348 ABLF02045642 GDIQ01023215 JAN71522.1 GDIP01145441 JAJ77961.1 GDIP01252174 GDIP01230534 GDIP01139447 GDIP01135351 LRGB01001005 JAL64267.1 KZS14107.1 GEZM01028452 JAV86299.1 GGMR01000850 MBY13469.1 GDIP01219287 JAJ04115.1 GEDC01016225 JAS21073.1 GDIP01221649 JAJ01753.1 GDIQ01224399 JAK27326.1 KK852427 KDR24110.1 GDIQ01239244 JAK12481.1 KQ434822 KZC07078.1 APGK01032099 KB740835 ENN78789.1 UFQT01000827 UFQT01002674 SSX27425.1 SSX33909.1 BT127607 AEE62569.1 APGK01005323 KB736187 ENN83244.1 GL732585 EFX74207.1 GDRN01066335 JAI64549.1 AFFK01020457 JH431580 KF621131 AHB33467.1 LJIG01009390 KRT83175.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000245037
+ More
UP000069940 UP000249989 UP000095301 UP000037069 UP000009192 UP000268350 UP000008820 UP000001819 UP000008744 UP000001070 UP000215335 UP000095300 UP000002320 UP000002358 UP000001292 UP000008711 UP000000803 UP000192221 UP000192223 UP000076408 UP000007801 UP000092443 UP000092460 UP000235965 UP000008792 UP000075884 UP000078200 UP000007266 UP000069272 UP000092445 UP000002282 UP000000673 UP000009046 UP000092553 UP000183832 UP000007798 UP000075920 UP000076407 UP000075903 UP000075902 UP000075885 UP000092462 UP000075886 UP000075880 UP000075881 UP000030765 UP000075882 UP000007062 UP000075840 UP000075883 UP000007819 UP000075900 UP000076858 UP000027135 UP000076502 UP000019118 UP000000305
UP000069940 UP000249989 UP000095301 UP000037069 UP000009192 UP000268350 UP000008820 UP000001819 UP000008744 UP000001070 UP000215335 UP000095300 UP000002320 UP000002358 UP000001292 UP000008711 UP000000803 UP000192221 UP000192223 UP000076408 UP000007801 UP000092443 UP000092460 UP000235965 UP000008792 UP000075884 UP000078200 UP000007266 UP000069272 UP000092445 UP000002282 UP000000673 UP000009046 UP000092553 UP000183832 UP000007798 UP000075920 UP000076407 UP000075903 UP000075902 UP000075885 UP000092462 UP000075886 UP000075880 UP000075881 UP000030765 UP000075882 UP000007062 UP000075840 UP000075883 UP000007819 UP000075900 UP000076858 UP000027135 UP000076502 UP000019118 UP000000305
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JJJ8
A0A2A4JJQ5
A0A194PT86
A0A194QZS2
A0A212EPJ9
A0A1B6KJV8
+ More
A0A023EWD6 A0A097PHN6 A0A1B6GHV6 A0A1L8EE66 A0A2P8Y706 A0A182H9M4 A0A1S4EZR6 T1PAU1 A0A0L0CF33 B4KMN2 A0A3B0J741 Q17KD5 Q293E0 B4GBU9 B4J4M7 A0A232FBI2 A0A1I8PBX8 W8C6G9 B0XGB7 K7IV31 T1D3W9 B4HP56 A0A0J9RAR7 B3NS73 Q7KKI0 A0A1W4ULS5 A0A1W4WN70 A0A182Y3I7 B3MH19 A0A2M4A6N5 A0A1A9XDF7 A0A1B0AML6 A0A2J7RAN2 B4LNH7 A0A182NU32 A0A1A9UK11 T1E783 D2A1F8 A0A2M4A6E1 A0A2M4BJH8 A0A182F140 A0A1B0AFU9 B4P5N3 W5J3N6 E0VSM9 A0A1Q3F539 A0A1Q3F4J5 A0A0A1X9V6 A0A0M3QVL0 A0A034W3H8 U5EUG1 A0A1J1IHK9 B4NMW6 A0A182W6S4 A0A182XL29 A0A182UUS1 A0A182U1C1 A0A182PCM4 A0A0K8TSU0 A0A2M4BKI2 A0A1B0D4S6 A0A1L8DUF7 A0A182Q8N6 A0A182J8P6 A0A182JXW2 A0A084W4S8 A0A182L112 Q7Q467 A0A182ICJ3 A0A0U2HUG4 A0A182M0Q1 X1WV13 A0A182R2Y6 A0A0P6H8C7 A0A0N8AI12 A0A0N8CEK0 A0A1Y1MKQ3 A0A2S2N8L1 A0A0P4ZYX3 A0A1B6D5X1 A0A0N7ZQT6 A0A0P5I5M7 A0A067RTG8 A0A0P5H4B5 A0A154P7A5 N6TEJ1 A0A336MAX3 J3JWI9 N6TRW5 E9H224 A0A0P4WBF7 T1IZH6 V5RDW2 A0A0T6B8L4
A0A023EWD6 A0A097PHN6 A0A1B6GHV6 A0A1L8EE66 A0A2P8Y706 A0A182H9M4 A0A1S4EZR6 T1PAU1 A0A0L0CF33 B4KMN2 A0A3B0J741 Q17KD5 Q293E0 B4GBU9 B4J4M7 A0A232FBI2 A0A1I8PBX8 W8C6G9 B0XGB7 K7IV31 T1D3W9 B4HP56 A0A0J9RAR7 B3NS73 Q7KKI0 A0A1W4ULS5 A0A1W4WN70 A0A182Y3I7 B3MH19 A0A2M4A6N5 A0A1A9XDF7 A0A1B0AML6 A0A2J7RAN2 B4LNH7 A0A182NU32 A0A1A9UK11 T1E783 D2A1F8 A0A2M4A6E1 A0A2M4BJH8 A0A182F140 A0A1B0AFU9 B4P5N3 W5J3N6 E0VSM9 A0A1Q3F539 A0A1Q3F4J5 A0A0A1X9V6 A0A0M3QVL0 A0A034W3H8 U5EUG1 A0A1J1IHK9 B4NMW6 A0A182W6S4 A0A182XL29 A0A182UUS1 A0A182U1C1 A0A182PCM4 A0A0K8TSU0 A0A2M4BKI2 A0A1B0D4S6 A0A1L8DUF7 A0A182Q8N6 A0A182J8P6 A0A182JXW2 A0A084W4S8 A0A182L112 Q7Q467 A0A182ICJ3 A0A0U2HUG4 A0A182M0Q1 X1WV13 A0A182R2Y6 A0A0P6H8C7 A0A0N8AI12 A0A0N8CEK0 A0A1Y1MKQ3 A0A2S2N8L1 A0A0P4ZYX3 A0A1B6D5X1 A0A0N7ZQT6 A0A0P5I5M7 A0A067RTG8 A0A0P5H4B5 A0A154P7A5 N6TEJ1 A0A336MAX3 J3JWI9 N6TRW5 E9H224 A0A0P4WBF7 T1IZH6 V5RDW2 A0A0T6B8L4
PDB
4B2T
E-value=0,
Score=1947
Ontologies
KEGG
GO
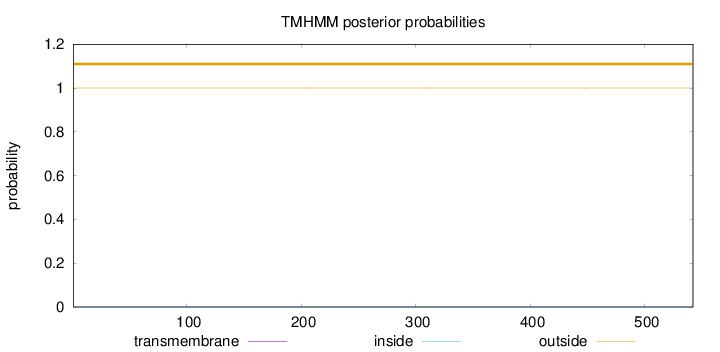
Topology
Subcellular location
Cytoplasm
Length:
542
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 542
Population Genetic Test Statistics
Pi
240.64167
Theta
172.982461
Tajima's D
2.094873
CLR
0.199008
CSRT
0.897705114744263
Interpretation
Uncertain
