Gene
KWMTBOMO00789
Pre Gene Modal
BGIBMGA009701
Annotation
PREDICTED:_cytochrome_c_oxidase_subunit_7C?_mitochondrial-like_[Plutella_xylostella]
Full name
Angiotensin-converting enzyme
Location in the cell
Mitochondrial Reliability : 2.006
Sequence
CDS
ATGATTACCCCACTGACTCGCATTTCAAACAAACTGGGTAGAAATGTGGTAACGAATTTCGTGAGAAACCATTCTAACGGCGGTATTCCTGGCGAGAATCTACCCTTCGACATTCACAACCGTTACAAGCTGACATTGTATTTCATCCTATATGCGGGCTCTGGACTATCAGCACCCTATCTCATCACCCGTCACCAGCTTCTGAAGAAGTAA
Protein
MITPLTRISNKLGRNVVTNFVRNHSNGGIPGENLPFDIHNRYKLTLYFILYAGSGLSAPYLITRHQLLKK
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M2 family.
Uniprot
H9JJK0
A0A194R123
I4DJ85
A0A2A4IV74
S4P8G0
A0A2A4JIM9
+ More
U5ETN7 A0A194R092 A0A2A4ITU1 A0A194PUU9 D6WSM8 A0A182TY41 A0A1J1I6G5 H9JJJ9 A0A0K8TPI3 A0A212EPN9 A0A182IL68 T1DSX0 A0A084VWX5 A0A1L8E024 A0A1B0GLN6 B0WNY0 T1D5Z1 A0A2M3Z0N0 A0A2M4A6L2 A0A2M4C5E8 W5JDK7 A0A182FFW0 A0A182P3L0 A0A182WIQ7 A0A1L8DZV0 A0A2J7R0L5 A0A1B0D137 A0A023EDR1 A0A182NHM4 A0A182KCV2 A0A182KRS4 A0A182X6D1 Q5TX26 A0A182I3N7 R4WE45 A0A182XZ83 A0A182RFM5 A0A067RGB5 Q1HQK5 A0A023EBA8 A0A182QC82 Q5MIQ0 A0A1I8Q2Z8 A0A2P8Z960 A0A0U2UEN0 A0A0L8HLJ1 A0A1L8EF51 A0A1I8MMK2 A0A1A9XAS6 A0A1B0BFK4 A0A1A9WNV1 A0A1L8EFA3 A0A1W0WT22 A0A0P5MVQ8 A0A164YUM6 A0A1L8EF58 A0A0P4WLH2 A0A0P5RYR8 B7S8I4 A0A2R7VWP9 E2AAJ7 A0A0P6DTU7 A0A0P6DTH1 E9H1K4 A0A2L2XYW5 A0A2R5LGQ2 B4LP41 Q1W2C6 B4KM37 B4MQX0 A0A0L0BMJ4 B4J5N3 B3MD26 A0A1D2N733 A0A0M5IXS4 A0A154NXS9 C9W1F2 L7M5F9 A0A3B0JK53 A0A1E1WZY8 B4QI79 A0A0B6YCB1 A0A0K8RF09 Q6B866 A0A023FQT8 A0A3B0IZQ4 Q290T6 B4GB97 A2TE22 B4HMB7 Q7JW00 B4NX37
U5ETN7 A0A194R092 A0A2A4ITU1 A0A194PUU9 D6WSM8 A0A182TY41 A0A1J1I6G5 H9JJJ9 A0A0K8TPI3 A0A212EPN9 A0A182IL68 T1DSX0 A0A084VWX5 A0A1L8E024 A0A1B0GLN6 B0WNY0 T1D5Z1 A0A2M3Z0N0 A0A2M4A6L2 A0A2M4C5E8 W5JDK7 A0A182FFW0 A0A182P3L0 A0A182WIQ7 A0A1L8DZV0 A0A2J7R0L5 A0A1B0D137 A0A023EDR1 A0A182NHM4 A0A182KCV2 A0A182KRS4 A0A182X6D1 Q5TX26 A0A182I3N7 R4WE45 A0A182XZ83 A0A182RFM5 A0A067RGB5 Q1HQK5 A0A023EBA8 A0A182QC82 Q5MIQ0 A0A1I8Q2Z8 A0A2P8Z960 A0A0U2UEN0 A0A0L8HLJ1 A0A1L8EF51 A0A1I8MMK2 A0A1A9XAS6 A0A1B0BFK4 A0A1A9WNV1 A0A1L8EFA3 A0A1W0WT22 A0A0P5MVQ8 A0A164YUM6 A0A1L8EF58 A0A0P4WLH2 A0A0P5RYR8 B7S8I4 A0A2R7VWP9 E2AAJ7 A0A0P6DTU7 A0A0P6DTH1 E9H1K4 A0A2L2XYW5 A0A2R5LGQ2 B4LP41 Q1W2C6 B4KM37 B4MQX0 A0A0L0BMJ4 B4J5N3 B3MD26 A0A1D2N733 A0A0M5IXS4 A0A154NXS9 C9W1F2 L7M5F9 A0A3B0JK53 A0A1E1WZY8 B4QI79 A0A0B6YCB1 A0A0K8RF09 Q6B866 A0A023FQT8 A0A3B0IZQ4 Q290T6 B4GB97 A2TE22 B4HMB7 Q7JW00 B4NX37
EC Number
3.4.-.-
Pubmed
19121390
26354079
22651552
23622113
18362917
19820115
+ More
26369729 22118469 24438588 24330624 20920257 23761445 24945155 20966253 12364791 14747013 17210077 23691247 25244985 24845553 17204158 17510324 17244540 29403074 26621068 25315136 20798317 21292972 26561354 17994087 18057021 26108605 27289101 20650005 24029695 25576852 28503490 16102420 15632085 17598752 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
26369729 22118469 24438588 24330624 20920257 23761445 24945155 20966253 12364791 14747013 17210077 23691247 25244985 24845553 17204158 17510324 17244540 29403074 26621068 25315136 20798317 21292972 26561354 17994087 18057021 26108605 27289101 20650005 24029695 25576852 28503490 16102420 15632085 17598752 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
BABH01017420
KQ460890
KPJ10950.1
AK401353
KQ459593
BAM17975.1
+ More
KPI96525.1 NWSH01006924 PCG63184.1 GAIX01005951 JAA86609.1 NWSH01001257 PCG71955.1 GANO01001825 JAB58046.1 KPJ10949.1 PCG63185.1 PCG71954.1 KPI96524.1 KQ971354 EFA07588.1 CVRI01000041 CRK95322.1 GDAI01001301 JAI16302.1 AGBW02013449 OWR43421.1 GAMD01001294 JAB00297.1 ATLV01017806 KE525192 KFB42469.1 GFDF01002053 JAV12031.1 AJWK01008032 DS232017 EDS31999.1 GALA01000313 JAA94539.1 GGFM01001338 MBW22089.1 GGFK01003059 MBW36380.1 GGFJ01011411 MBW60552.1 ADMH02001692 ETN61408.1 GFDF01002054 JAV12030.1 NEVH01008227 PNF34381.1 AJVK01010180 GAPW01006874 JAC06724.1 AAAB01008807 EAL41744.1 EDO64527.1 EDO64528.1 APCN01000215 AK418089 BAN21304.1 KK852657 KDR19224.1 DQ440439 CH477771 CH477251 ABF18472.1 EAT36395.1 EAT46042.1 EJY57433.1 GAPW01006875 GAPW01006873 GEHC01000228 JAC06723.1 JAV47417.1 AXCN02000593 AY826160 AAV90732.1 PYGN01000142 PSN53028.1 KT754337 KT754338 ALS04171.1 KQ417847 KOF90082.1 GFDG01001442 JAV17357.1 JXJN01013537 GFDG01001443 JAV17356.1 MTYJ01000050 OQV18342.1 GDIQ01152847 JAK98878.1 LRGB01000868 KZS15623.1 GFDG01001449 JAV17350.1 GDIP01255247 JAI68154.1 GDIQ01094665 JAL57061.1 EF710646 ACE75209.1 KK854137 PTY11934.1 GL438128 EFN69544.1 GDIQ01073025 JAN21712.1 GDIQ01073024 JAN21713.1 GL732583 EFX74371.1 IAAA01003746 IAAA01003747 LAA01142.1 GGLE01004523 MBY08649.1 CH940648 EDW62235.1 DQ445501 ABD98739.1 CH933808 EDW08701.1 KRG04232.1 CH963849 EDW74509.1 JRES01001644 KNC21232.1 CH916367 EDW01809.1 CH902619 EDV36341.1 LJIJ01000174 ODN01057.1 CP012524 ALC41986.1 KQ434781 KZC04485.1 EZ406100 ACX53899.1 GACK01006636 JAA58398.1 OUUW01000001 SPP73626.1 GFAC01006598 JAT92590.1 CM000362 EDX06439.1 HACG01006265 CEK53130.1 GADI01004061 JAA69747.1 AY674282 AAT92215.1 GBBK01001409 GBBK01001408 JAC23073.1 SPP73627.1 CM000071 EAL25276.1 CH479181 EDW32199.1 EF189578 EF189579 EF189580 EF189581 EF189582 EF189583 EF189584 EF189585 CM002911 ABM88356.1 ABM88357.1 ABM88358.1 ABM88359.1 ABM88360.1 ABM88361.1 ABM88362.1 ABM88363.1 KMY92656.1 KMY92657.1 CH480816 CH481236 EDW47194.1 EDW52799.1 AE013599 AY118515 AAF58857.2 AAM49884.1 AAO41466.1 AHN56066.1 AHN56067.1 CM000157 EDW89598.1 KRJ98561.1
KPI96525.1 NWSH01006924 PCG63184.1 GAIX01005951 JAA86609.1 NWSH01001257 PCG71955.1 GANO01001825 JAB58046.1 KPJ10949.1 PCG63185.1 PCG71954.1 KPI96524.1 KQ971354 EFA07588.1 CVRI01000041 CRK95322.1 GDAI01001301 JAI16302.1 AGBW02013449 OWR43421.1 GAMD01001294 JAB00297.1 ATLV01017806 KE525192 KFB42469.1 GFDF01002053 JAV12031.1 AJWK01008032 DS232017 EDS31999.1 GALA01000313 JAA94539.1 GGFM01001338 MBW22089.1 GGFK01003059 MBW36380.1 GGFJ01011411 MBW60552.1 ADMH02001692 ETN61408.1 GFDF01002054 JAV12030.1 NEVH01008227 PNF34381.1 AJVK01010180 GAPW01006874 JAC06724.1 AAAB01008807 EAL41744.1 EDO64527.1 EDO64528.1 APCN01000215 AK418089 BAN21304.1 KK852657 KDR19224.1 DQ440439 CH477771 CH477251 ABF18472.1 EAT36395.1 EAT46042.1 EJY57433.1 GAPW01006875 GAPW01006873 GEHC01000228 JAC06723.1 JAV47417.1 AXCN02000593 AY826160 AAV90732.1 PYGN01000142 PSN53028.1 KT754337 KT754338 ALS04171.1 KQ417847 KOF90082.1 GFDG01001442 JAV17357.1 JXJN01013537 GFDG01001443 JAV17356.1 MTYJ01000050 OQV18342.1 GDIQ01152847 JAK98878.1 LRGB01000868 KZS15623.1 GFDG01001449 JAV17350.1 GDIP01255247 JAI68154.1 GDIQ01094665 JAL57061.1 EF710646 ACE75209.1 KK854137 PTY11934.1 GL438128 EFN69544.1 GDIQ01073025 JAN21712.1 GDIQ01073024 JAN21713.1 GL732583 EFX74371.1 IAAA01003746 IAAA01003747 LAA01142.1 GGLE01004523 MBY08649.1 CH940648 EDW62235.1 DQ445501 ABD98739.1 CH933808 EDW08701.1 KRG04232.1 CH963849 EDW74509.1 JRES01001644 KNC21232.1 CH916367 EDW01809.1 CH902619 EDV36341.1 LJIJ01000174 ODN01057.1 CP012524 ALC41986.1 KQ434781 KZC04485.1 EZ406100 ACX53899.1 GACK01006636 JAA58398.1 OUUW01000001 SPP73626.1 GFAC01006598 JAT92590.1 CM000362 EDX06439.1 HACG01006265 CEK53130.1 GADI01004061 JAA69747.1 AY674282 AAT92215.1 GBBK01001409 GBBK01001408 JAC23073.1 SPP73627.1 CM000071 EAL25276.1 CH479181 EDW32199.1 EF189578 EF189579 EF189580 EF189581 EF189582 EF189583 EF189584 EF189585 CM002911 ABM88356.1 ABM88357.1 ABM88358.1 ABM88359.1 ABM88360.1 ABM88361.1 ABM88362.1 ABM88363.1 KMY92656.1 KMY92657.1 CH480816 CH481236 EDW47194.1 EDW52799.1 AE013599 AY118515 AAF58857.2 AAM49884.1 AAO41466.1 AHN56066.1 AHN56067.1 CM000157 EDW89598.1 KRJ98561.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007266
UP000075902
+ More
UP000183832 UP000007151 UP000075880 UP000030765 UP000092461 UP000002320 UP000000673 UP000069272 UP000075885 UP000075920 UP000235965 UP000092462 UP000075884 UP000075881 UP000075882 UP000076407 UP000007062 UP000075840 UP000076408 UP000075900 UP000027135 UP000008820 UP000075886 UP000095300 UP000245037 UP000053454 UP000095301 UP000092443 UP000092460 UP000091820 UP000076858 UP000000311 UP000000305 UP000008792 UP000009192 UP000007798 UP000037069 UP000001070 UP000007801 UP000094527 UP000092553 UP000076502 UP000268350 UP000000304 UP000001819 UP000008744 UP000001292 UP000000803 UP000002282
UP000183832 UP000007151 UP000075880 UP000030765 UP000092461 UP000002320 UP000000673 UP000069272 UP000075885 UP000075920 UP000235965 UP000092462 UP000075884 UP000075881 UP000075882 UP000076407 UP000007062 UP000075840 UP000076408 UP000075900 UP000027135 UP000008820 UP000075886 UP000095300 UP000245037 UP000053454 UP000095301 UP000092443 UP000092460 UP000091820 UP000076858 UP000000311 UP000000305 UP000008792 UP000009192 UP000007798 UP000037069 UP000001070 UP000007801 UP000094527 UP000092553 UP000076502 UP000268350 UP000000304 UP000001819 UP000008744 UP000001292 UP000000803 UP000002282
Interpro
Gene 3D
ProteinModelPortal
H9JJK0
A0A194R123
I4DJ85
A0A2A4IV74
S4P8G0
A0A2A4JIM9
+ More
U5ETN7 A0A194R092 A0A2A4ITU1 A0A194PUU9 D6WSM8 A0A182TY41 A0A1J1I6G5 H9JJJ9 A0A0K8TPI3 A0A212EPN9 A0A182IL68 T1DSX0 A0A084VWX5 A0A1L8E024 A0A1B0GLN6 B0WNY0 T1D5Z1 A0A2M3Z0N0 A0A2M4A6L2 A0A2M4C5E8 W5JDK7 A0A182FFW0 A0A182P3L0 A0A182WIQ7 A0A1L8DZV0 A0A2J7R0L5 A0A1B0D137 A0A023EDR1 A0A182NHM4 A0A182KCV2 A0A182KRS4 A0A182X6D1 Q5TX26 A0A182I3N7 R4WE45 A0A182XZ83 A0A182RFM5 A0A067RGB5 Q1HQK5 A0A023EBA8 A0A182QC82 Q5MIQ0 A0A1I8Q2Z8 A0A2P8Z960 A0A0U2UEN0 A0A0L8HLJ1 A0A1L8EF51 A0A1I8MMK2 A0A1A9XAS6 A0A1B0BFK4 A0A1A9WNV1 A0A1L8EFA3 A0A1W0WT22 A0A0P5MVQ8 A0A164YUM6 A0A1L8EF58 A0A0P4WLH2 A0A0P5RYR8 B7S8I4 A0A2R7VWP9 E2AAJ7 A0A0P6DTU7 A0A0P6DTH1 E9H1K4 A0A2L2XYW5 A0A2R5LGQ2 B4LP41 Q1W2C6 B4KM37 B4MQX0 A0A0L0BMJ4 B4J5N3 B3MD26 A0A1D2N733 A0A0M5IXS4 A0A154NXS9 C9W1F2 L7M5F9 A0A3B0JK53 A0A1E1WZY8 B4QI79 A0A0B6YCB1 A0A0K8RF09 Q6B866 A0A023FQT8 A0A3B0IZQ4 Q290T6 B4GB97 A2TE22 B4HMB7 Q7JW00 B4NX37
U5ETN7 A0A194R092 A0A2A4ITU1 A0A194PUU9 D6WSM8 A0A182TY41 A0A1J1I6G5 H9JJJ9 A0A0K8TPI3 A0A212EPN9 A0A182IL68 T1DSX0 A0A084VWX5 A0A1L8E024 A0A1B0GLN6 B0WNY0 T1D5Z1 A0A2M3Z0N0 A0A2M4A6L2 A0A2M4C5E8 W5JDK7 A0A182FFW0 A0A182P3L0 A0A182WIQ7 A0A1L8DZV0 A0A2J7R0L5 A0A1B0D137 A0A023EDR1 A0A182NHM4 A0A182KCV2 A0A182KRS4 A0A182X6D1 Q5TX26 A0A182I3N7 R4WE45 A0A182XZ83 A0A182RFM5 A0A067RGB5 Q1HQK5 A0A023EBA8 A0A182QC82 Q5MIQ0 A0A1I8Q2Z8 A0A2P8Z960 A0A0U2UEN0 A0A0L8HLJ1 A0A1L8EF51 A0A1I8MMK2 A0A1A9XAS6 A0A1B0BFK4 A0A1A9WNV1 A0A1L8EFA3 A0A1W0WT22 A0A0P5MVQ8 A0A164YUM6 A0A1L8EF58 A0A0P4WLH2 A0A0P5RYR8 B7S8I4 A0A2R7VWP9 E2AAJ7 A0A0P6DTU7 A0A0P6DTH1 E9H1K4 A0A2L2XYW5 A0A2R5LGQ2 B4LP41 Q1W2C6 B4KM37 B4MQX0 A0A0L0BMJ4 B4J5N3 B3MD26 A0A1D2N733 A0A0M5IXS4 A0A154NXS9 C9W1F2 L7M5F9 A0A3B0JK53 A0A1E1WZY8 B4QI79 A0A0B6YCB1 A0A0K8RF09 Q6B866 A0A023FQT8 A0A3B0IZQ4 Q290T6 B4GB97 A2TE22 B4HMB7 Q7JW00 B4NX37
PDB
2Y69
E-value=4.07491e-08,
Score=131
Ontologies
PATHWAY
GO
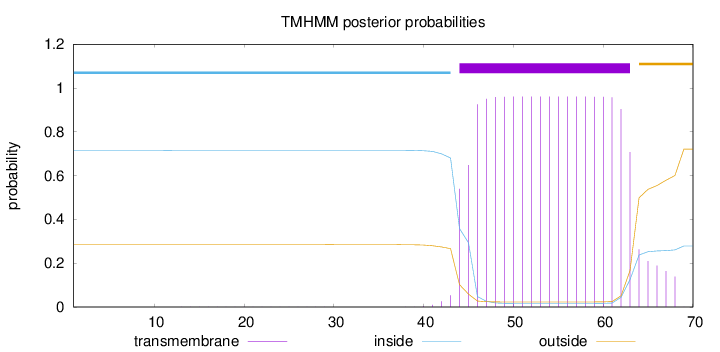
Topology
Length:
70
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.17687
Exp number, first 60 AAs:
15.64231
Total prob of N-in:
0.71538
POSSIBLE N-term signal
sequence
inside
1 - 43
TMhelix
44 - 63
outside
64 - 70
Population Genetic Test Statistics
Pi
445.801791
Theta
181.037417
Tajima's D
4.694829
CLR
0.000406
CSRT
0.99990000499975
Interpretation
Uncertain
