Gene
KWMTBOMO00788
Pre Gene Modal
BGIBMGA009702
Annotation
acetylglucosaminyltransferase_[Bombyx_mori]
Full name
Alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase
Alternative Name
N-glycosyl-oligosaccharide-glycoprotein N-acetylglucosaminyltransferase I
Location in the cell
Mitochondrial Reliability : 1.775 Nuclear Reliability : 1.296
Sequence
CDS
ATGCGCCTGAATGTAAGAAAACTATTGTTTGTTAGTTGCGGTGTTTTATTGTTGTGGTTCATCGTCTCTTACTCGTCGATTATCGAGTTGCCGAGGTCTATAACCTCTAAGGACACAGCAGCCGACGTAGAAAACAAATTAGGCAGACTTCAGAAGAAAATTGATGACCAAATGCTGGAAAGCAAAACACTGTTGCAGAAAGTGAAAAATGAATTGAAAAAAAAAACGCAATCAAAGGAGTCTTTAATTCAGTACTCTGATGAAATTATTGATGGTCCAACAATACTACCGGTGCTTGTGATCGCTTGCGACCGGGTCACAGTTAAAAGGTGTCTCGACAATCTAGTCAAGTTCAGACCGGACAAGGAAAGGTTTCCGATCGTAGTAAGCCAAGATTGCGGTCACAACGCCACTTACCAAGTGATACGCAGCTTCACCGATGCGGATCCGTCGATAAAGGTCATACTGCAACCGGATTTGTCCGAGATACCGCTTCCGAAGGTCAAGGTCAAGCTGAAAGGCTACTACAAGATCGCAAGGCATTTCAAATTCGCCTTAAATCACATGTTCAATACGTTGAATCACGAGGCTGTGATAATTGTTGAGGATGACCTGGACATTTCGCCGGATTTCTACGAATATTTCTTGGGCACGTATCCTTTACTGCAAAAGGATCCTAGTTTATGGTGCATATCGGCGTGGAATGACAACGGGAAGAAAGACATCATAGACCTGACCAGGCCGGAACTGTTGTACAGGACGGACTTCTTCCCGGGCCTCGGGTGGATGCTCAGGAAGGAGATGTGGACCAGACTCGAGCCGATATGGCCTCCGGCATTCTTCGACGACTGGCTCCGCAATCCAGTCAACACCCAAGGTCGAGCCTGCATCAGACCGGAGATATCCAGGACGTATTCGTTTGGGAAGATCGGCGTGAGTCAAGGACTGTTCTTCGATAAGCACCTTAGGAAAATACAATTGAATCTGGATTACGTCGAGTTCACTAAATTAAACTTAACTTATTTATTAAAGGATAATTACGACGAGACTCTAACCTCTACGGTGTACTCCTTGCCGGAGGTGATGGCCGAGGATGTGATACTTGGCGAGATGGACTCCGCAGTAAGGGTCTCTTACTCCAATGCCAAGAGCTACCAGCGGGCCGCCAAGAAACTTGGCCTCATGGACGACTTTAGAAGCGGCGTCCCCAGGACCGCGTACAGAGGTATAGTGACGTGCTTTATTCGAGGTCGACGTGTTTATCTGGCCCCGGACTACCAGTGGACCAAGTACGACCCCTCGTGGGGCTAG
Protein
MRLNVRKLLFVSCGVLLLWFIVSYSSIIELPRSITSKDTAADVENKLGRLQKKIDDQMLESKTLLQKVKNELKKKTQSKESLIQYSDEIIDGPTILPVLVIACDRVTVKRCLDNLVKFRPDKERFPIVVSQDCGHNATYQVIRSFTDADPSIKVILQPDLSEIPLPKVKVKLKGYYKIARHFKFALNHMFNTLNHEAVIIVEDDLDISPDFYEYFLGTYPLLQKDPSLWCISAWNDNGKKDIIDLTRPELLYRTDFFPGLGWMLRKEMWTRLEPIWPPAFFDDWLRNPVNTQGRACIRPEISRTYSFGKIGVSQGLFFDKHLRKIQLNLDYVEFTKLNLTYLLKDNYDETLTSTVYSLPEVMAEDVILGEMDSAVRVSYSNAKSYQRAAKKLGLMDDFRSGVPRTAYRGIVTCFIRGRRVYLAPDYQWTKYDPSWG
Summary
Description
Initiates complex N-linked carbohydrate formation. Essential for the conversion of high-mannose to hybrid and complex N-glycans.
Catalytic Activity
N(4)-(alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man-(1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc)-L-asparaginyl-[protein] (isomer 5A1,2) + UDP-N-acetyl-alpha-D-glucosamine = H(+) + N(4)-{beta-D-GlcNAc-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->3)-[alpha-D-Man-(1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc}-L-asparaginyl-[protein] + UDP
Cofactor
Mn(2+)
Subunit
Interacts with MGAT4D.
Similarity
Belongs to the glycosyltransferase 13 family.
Keywords
Complete proteome
Disulfide bond
Glycosyltransferase
Golgi apparatus
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Polymorphism
Feature
chain Alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase
sequence variant In dbSNP:rs7726005.
sequence variant In dbSNP:rs7726005.
Uniprot
F8WLR8
H9JJK1
H2BEB9
A0A194R0U9
A0A194PT25
A0A2H1VI14
+ More
A0A212EX38 F7EGJ8 H3AN27 A0A2K6NLC0 A0A2J7PD74 A0A067QSE6 N6T3H9 Q924C0 P70680 Q924B9 U4UI41 V8NFG1 A0A1W7RJT8 Q924B8 Q544F0 P27808 A0A098M1Y9 J3S955 A0A1S3D9R7 Q8CIC9 T1E6R7 G3HK16 Q09325 I3N332 Q78E58 A0A087TMU3 A0A3B3WR82 A0A023F2K8 A0A3B4D982 A0A2A3EL29 A0A224XCI7 A0A0B8RV60 A0A0K8S2V2 A0A1A9ZYC3 A0A087X9E1 A0A1Y1L8B4 A0A1B0G3G9 A0A0L7R0U9 A0A315VXC4 A0A2I0T0W5 A0A1B0AXC5 A0A3B3VNM7 I3JAS7 A0A151M5X7 A0A3P8YP14 A0A1A9XVF6 A0A3B5L1Z1 A0A1U7SJ06 A0A2D0SWU1 A0A0A9W2X8 L5JVU8 A0A383ZEF0 G1KL90 A0A0A9X981 W5UBY9 A0A1I8MXJ3 A0A0L0BTQ1 A0A2F0B5I0 E2BDY8 A0A232EL13 A0A087ZQX0 A0A3B1JRJ3 A0A2K5XAV2 A0A2K5L3S5 Q0KKC2 A0A2J8JTB6 A0A091UZ47 A0A2R8Z919 K7CRV2 M4AFU5 A0A2K6AUI7 B3NJQ5 K7BGV7 A0A195D0W8 G7MUK4 A0A0M8ZVJ3 A0A1W4V4T8 G3R2D6 A0A310SC60 A0A3B5A790 K7J882 A0A158NYY3 A0A1A9V2E7 Q59G70 A0A3B0J032 A0A026W650 B4LIP0 A0A3P9N5Y9 P26572 A0A1I8P9G2 A0A091GC10 D2I5F9 B3KQ18
A0A212EX38 F7EGJ8 H3AN27 A0A2K6NLC0 A0A2J7PD74 A0A067QSE6 N6T3H9 Q924C0 P70680 Q924B9 U4UI41 V8NFG1 A0A1W7RJT8 Q924B8 Q544F0 P27808 A0A098M1Y9 J3S955 A0A1S3D9R7 Q8CIC9 T1E6R7 G3HK16 Q09325 I3N332 Q78E58 A0A087TMU3 A0A3B3WR82 A0A023F2K8 A0A3B4D982 A0A2A3EL29 A0A224XCI7 A0A0B8RV60 A0A0K8S2V2 A0A1A9ZYC3 A0A087X9E1 A0A1Y1L8B4 A0A1B0G3G9 A0A0L7R0U9 A0A315VXC4 A0A2I0T0W5 A0A1B0AXC5 A0A3B3VNM7 I3JAS7 A0A151M5X7 A0A3P8YP14 A0A1A9XVF6 A0A3B5L1Z1 A0A1U7SJ06 A0A2D0SWU1 A0A0A9W2X8 L5JVU8 A0A383ZEF0 G1KL90 A0A0A9X981 W5UBY9 A0A1I8MXJ3 A0A0L0BTQ1 A0A2F0B5I0 E2BDY8 A0A232EL13 A0A087ZQX0 A0A3B1JRJ3 A0A2K5XAV2 A0A2K5L3S5 Q0KKC2 A0A2J8JTB6 A0A091UZ47 A0A2R8Z919 K7CRV2 M4AFU5 A0A2K6AUI7 B3NJQ5 K7BGV7 A0A195D0W8 G7MUK4 A0A0M8ZVJ3 A0A1W4V4T8 G3R2D6 A0A310SC60 A0A3B5A790 K7J882 A0A158NYY3 A0A1A9V2E7 Q59G70 A0A3B0J032 A0A026W650 B4LIP0 A0A3P9N5Y9 P26572 A0A1I8P9G2 A0A091GC10 D2I5F9 B3KQ18
EC Number
2.4.1.101
Pubmed
19121390
22238347
26354079
22118469
17495919
9215903
+ More
25362486 24845553 23537049 11467936 8910379 24297900 26358130 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 1533386 1421759 16141072 15489334 20805325 23025625 23758969 25727380 21804562 7764514 25474469 25476704 28004739 29703783 25186727 22293439 25069045 25401762 26823975 23258410 23127152 25315136 26108605 20798317 28648823 25329095 16136131 22722832 23542700 17994087 17431167 22002653 25319552 22398555 20075255 21347285 24508170 30249741 1702225 1827260 14702039 15372022 20378551 24275569 20010809
25362486 24845553 23537049 11467936 8910379 24297900 26358130 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 1533386 1421759 16141072 15489334 20805325 23025625 23758969 25727380 21804562 7764514 25474469 25476704 28004739 29703783 25186727 22293439 25069045 25401762 26823975 23258410 23127152 25315136 26108605 20798317 28648823 25329095 16136131 22722832 23542700 17994087 17431167 22002653 25319552 22398555 20075255 21347285 24508170 30249741 1702225 1827260 14702039 15372022 20378551 24275569 20010809
EMBL
AB587677
BAK55694.1
BABH01017414
BABH01017415
HQ888863
AEX00082.1
+ More
KQ460890 KPJ10875.1 KQ459593 KPI96467.1 ODYU01002648 SOQ40431.1 AGBW02011830 OWR46062.1 AFYH01090827 NEVH01026393 PNF14283.1 KK853011 KDR12543.1 APGK01045744 APGK01045745 APGK01045746 KB741037 ENN74704.1 AF343963 AAK61868.1 U65791 AAC52872.1 AF343964 AAK61869.1 KB632173 ERL89550.1 AZIM01004468 ETE60706.1 GDAY02000016 JAV51397.1 AF343965 AAK61870.1 AK004760 AK040121 AK155322 CH466575 BAB23541.1 BAC30515.1 BAE33189.1 EDL33761.1 M73491 L07037 AK087959 BC006629 GBSI01000109 JAC96386.1 JU175199 GBEX01000024 AFJ50723.1 JAI14536.1 BC031752 AAH31752.1 GAAZ01000010 GBKC01000036 JAA97933.1 JAG46034.1 JH000448 EGV96592.1 D16302 BC074010 AGTP01110808 AGTP01110809 U65792 AAC52873.1 KK115950 KFM66432.1 GBBI01003162 JAC15550.1 KZ288226 PBC31896.1 GFTR01006280 JAW10146.1 GBSH01000148 JAG68876.1 GBKD01000011 JAG47607.1 AYCK01017180 GEZM01063649 JAV69041.1 CCAG010013348 KQ414670 KOC64421.1 NHOQ01000885 PWA28221.1 KZ527314 PKU27437.1 JXJN01005221 AERX01051254 AKHW03006526 KYO19830.1 GBHO01041828 GBHO01037557 GBHO01037554 GBHO01037550 GBRD01008547 GBRD01003590 GDHC01001738 JAG01776.1 JAG06047.1 JAG06050.1 JAG06054.1 JAG57274.1 JAQ16891.1 KB031114 ELK03187.1 GBHO01041835 GBHO01041831 GBHO01029979 GBHO01029978 GBRD01008548 GBRD01003589 GDHC01018788 JAG01769.1 JAG01773.1 JAG13625.1 JAG13626.1 JAG57273.1 JAP99840.1 JT405492 AHH37207.1 JRES01001351 KNC23460.1 NTJE010077542 MBV98266.1 GL447693 EFN86114.1 NNAY01003677 OXU19011.1 AEMK02000010 AB251342 BAF03221.1 AACZ04045833 NBAG03000426 PNI26010.1 PNI26011.1 PNI26013.1 PNI26016.1 PNI26019.1 KL410349 KFQ96274.1 AJFE02018931 GABE01009438 GABE01009437 GABE01009436 GABE01009435 JAA35303.1 CH954179 EDV55324.1 GABC01005459 GABC01005458 GABC01005457 GABC01005456 GABF01005459 GABF01005458 GABF01005457 GABF01005456 GABD01000081 GABD01000080 GABD01000079 GABD01000078 JAA05882.1 JAA16686.1 JAA33019.1 KQ977004 KYN06501.1 JSUE03037723 JU334560 JU475645 JU475646 JU475647 JU475648 JV047094 JV047095 CM001258 AFE78313.1 AFH32449.1 AFI37165.1 EHH27126.1 KQ435826 KOX72005.1 CABD030042842 KQ769812 OAD52833.1 ADTU01004399 AB209239 BAD92476.1 OUUW01000001 SPP73847.1 KK107419 QOIP01000003 EZA51086.1 RLU24583.1 CH940648 EDW61393.1 M55621 M61829 AK092256 AK094130 AK290769 CR456861 AC022413 CH471165 BC003575 KL447932 KFO78846.1 ACTA01187874 GL194665 EFB19613.1 AK057194 BAG51880.1
KQ460890 KPJ10875.1 KQ459593 KPI96467.1 ODYU01002648 SOQ40431.1 AGBW02011830 OWR46062.1 AFYH01090827 NEVH01026393 PNF14283.1 KK853011 KDR12543.1 APGK01045744 APGK01045745 APGK01045746 KB741037 ENN74704.1 AF343963 AAK61868.1 U65791 AAC52872.1 AF343964 AAK61869.1 KB632173 ERL89550.1 AZIM01004468 ETE60706.1 GDAY02000016 JAV51397.1 AF343965 AAK61870.1 AK004760 AK040121 AK155322 CH466575 BAB23541.1 BAC30515.1 BAE33189.1 EDL33761.1 M73491 L07037 AK087959 BC006629 GBSI01000109 JAC96386.1 JU175199 GBEX01000024 AFJ50723.1 JAI14536.1 BC031752 AAH31752.1 GAAZ01000010 GBKC01000036 JAA97933.1 JAG46034.1 JH000448 EGV96592.1 D16302 BC074010 AGTP01110808 AGTP01110809 U65792 AAC52873.1 KK115950 KFM66432.1 GBBI01003162 JAC15550.1 KZ288226 PBC31896.1 GFTR01006280 JAW10146.1 GBSH01000148 JAG68876.1 GBKD01000011 JAG47607.1 AYCK01017180 GEZM01063649 JAV69041.1 CCAG010013348 KQ414670 KOC64421.1 NHOQ01000885 PWA28221.1 KZ527314 PKU27437.1 JXJN01005221 AERX01051254 AKHW03006526 KYO19830.1 GBHO01041828 GBHO01037557 GBHO01037554 GBHO01037550 GBRD01008547 GBRD01003590 GDHC01001738 JAG01776.1 JAG06047.1 JAG06050.1 JAG06054.1 JAG57274.1 JAQ16891.1 KB031114 ELK03187.1 GBHO01041835 GBHO01041831 GBHO01029979 GBHO01029978 GBRD01008548 GBRD01003589 GDHC01018788 JAG01769.1 JAG01773.1 JAG13625.1 JAG13626.1 JAG57273.1 JAP99840.1 JT405492 AHH37207.1 JRES01001351 KNC23460.1 NTJE010077542 MBV98266.1 GL447693 EFN86114.1 NNAY01003677 OXU19011.1 AEMK02000010 AB251342 BAF03221.1 AACZ04045833 NBAG03000426 PNI26010.1 PNI26011.1 PNI26013.1 PNI26016.1 PNI26019.1 KL410349 KFQ96274.1 AJFE02018931 GABE01009438 GABE01009437 GABE01009436 GABE01009435 JAA35303.1 CH954179 EDV55324.1 GABC01005459 GABC01005458 GABC01005457 GABC01005456 GABF01005459 GABF01005458 GABF01005457 GABF01005456 GABD01000081 GABD01000080 GABD01000079 GABD01000078 JAA05882.1 JAA16686.1 JAA33019.1 KQ977004 KYN06501.1 JSUE03037723 JU334560 JU475645 JU475646 JU475647 JU475648 JV047094 JV047095 CM001258 AFE78313.1 AFH32449.1 AFI37165.1 EHH27126.1 KQ435826 KOX72005.1 CABD030042842 KQ769812 OAD52833.1 ADTU01004399 AB209239 BAD92476.1 OUUW01000001 SPP73847.1 KK107419 QOIP01000003 EZA51086.1 RLU24583.1 CH940648 EDW61393.1 M55621 M61829 AK092256 AK094130 AK290769 CR456861 AC022413 CH471165 BC003575 KL447932 KFO78846.1 ACTA01187874 GL194665 EFB19613.1 AK057194 BAG51880.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000002280
UP000008672
+ More
UP000233200 UP000235965 UP000027135 UP000019118 UP000030742 UP000000589 UP000079169 UP000001075 UP000002494 UP000005215 UP000054359 UP000261480 UP000261440 UP000242457 UP000092445 UP000028760 UP000092444 UP000053825 UP000092460 UP000261500 UP000005207 UP000050525 UP000265140 UP000092443 UP000261380 UP000189705 UP000221080 UP000010552 UP000261681 UP000001646 UP000095301 UP000037069 UP000008237 UP000215335 UP000005203 UP000018467 UP000233140 UP000233060 UP000008227 UP000002277 UP000053283 UP000240080 UP000002852 UP000233120 UP000008711 UP000078542 UP000006718 UP000053105 UP000192221 UP000001519 UP000261400 UP000002358 UP000005205 UP000078200 UP000268350 UP000053097 UP000279307 UP000008792 UP000242638 UP000005640 UP000095300 UP000053760 UP000008912
UP000233200 UP000235965 UP000027135 UP000019118 UP000030742 UP000000589 UP000079169 UP000001075 UP000002494 UP000005215 UP000054359 UP000261480 UP000261440 UP000242457 UP000092445 UP000028760 UP000092444 UP000053825 UP000092460 UP000261500 UP000005207 UP000050525 UP000265140 UP000092443 UP000261380 UP000189705 UP000221080 UP000010552 UP000261681 UP000001646 UP000095301 UP000037069 UP000008237 UP000215335 UP000005203 UP000018467 UP000233140 UP000233060 UP000008227 UP000002277 UP000053283 UP000240080 UP000002852 UP000233120 UP000008711 UP000078542 UP000006718 UP000053105 UP000192221 UP000001519 UP000261400 UP000002358 UP000005205 UP000078200 UP000268350 UP000053097 UP000279307 UP000008792 UP000242638 UP000005640 UP000095300 UP000053760 UP000008912
PRIDE
Pfam
PF03071 GNT-I
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
F8WLR8
H9JJK1
H2BEB9
A0A194R0U9
A0A194PT25
A0A2H1VI14
+ More
A0A212EX38 F7EGJ8 H3AN27 A0A2K6NLC0 A0A2J7PD74 A0A067QSE6 N6T3H9 Q924C0 P70680 Q924B9 U4UI41 V8NFG1 A0A1W7RJT8 Q924B8 Q544F0 P27808 A0A098M1Y9 J3S955 A0A1S3D9R7 Q8CIC9 T1E6R7 G3HK16 Q09325 I3N332 Q78E58 A0A087TMU3 A0A3B3WR82 A0A023F2K8 A0A3B4D982 A0A2A3EL29 A0A224XCI7 A0A0B8RV60 A0A0K8S2V2 A0A1A9ZYC3 A0A087X9E1 A0A1Y1L8B4 A0A1B0G3G9 A0A0L7R0U9 A0A315VXC4 A0A2I0T0W5 A0A1B0AXC5 A0A3B3VNM7 I3JAS7 A0A151M5X7 A0A3P8YP14 A0A1A9XVF6 A0A3B5L1Z1 A0A1U7SJ06 A0A2D0SWU1 A0A0A9W2X8 L5JVU8 A0A383ZEF0 G1KL90 A0A0A9X981 W5UBY9 A0A1I8MXJ3 A0A0L0BTQ1 A0A2F0B5I0 E2BDY8 A0A232EL13 A0A087ZQX0 A0A3B1JRJ3 A0A2K5XAV2 A0A2K5L3S5 Q0KKC2 A0A2J8JTB6 A0A091UZ47 A0A2R8Z919 K7CRV2 M4AFU5 A0A2K6AUI7 B3NJQ5 K7BGV7 A0A195D0W8 G7MUK4 A0A0M8ZVJ3 A0A1W4V4T8 G3R2D6 A0A310SC60 A0A3B5A790 K7J882 A0A158NYY3 A0A1A9V2E7 Q59G70 A0A3B0J032 A0A026W650 B4LIP0 A0A3P9N5Y9 P26572 A0A1I8P9G2 A0A091GC10 D2I5F9 B3KQ18
A0A212EX38 F7EGJ8 H3AN27 A0A2K6NLC0 A0A2J7PD74 A0A067QSE6 N6T3H9 Q924C0 P70680 Q924B9 U4UI41 V8NFG1 A0A1W7RJT8 Q924B8 Q544F0 P27808 A0A098M1Y9 J3S955 A0A1S3D9R7 Q8CIC9 T1E6R7 G3HK16 Q09325 I3N332 Q78E58 A0A087TMU3 A0A3B3WR82 A0A023F2K8 A0A3B4D982 A0A2A3EL29 A0A224XCI7 A0A0B8RV60 A0A0K8S2V2 A0A1A9ZYC3 A0A087X9E1 A0A1Y1L8B4 A0A1B0G3G9 A0A0L7R0U9 A0A315VXC4 A0A2I0T0W5 A0A1B0AXC5 A0A3B3VNM7 I3JAS7 A0A151M5X7 A0A3P8YP14 A0A1A9XVF6 A0A3B5L1Z1 A0A1U7SJ06 A0A2D0SWU1 A0A0A9W2X8 L5JVU8 A0A383ZEF0 G1KL90 A0A0A9X981 W5UBY9 A0A1I8MXJ3 A0A0L0BTQ1 A0A2F0B5I0 E2BDY8 A0A232EL13 A0A087ZQX0 A0A3B1JRJ3 A0A2K5XAV2 A0A2K5L3S5 Q0KKC2 A0A2J8JTB6 A0A091UZ47 A0A2R8Z919 K7CRV2 M4AFU5 A0A2K6AUI7 B3NJQ5 K7BGV7 A0A195D0W8 G7MUK4 A0A0M8ZVJ3 A0A1W4V4T8 G3R2D6 A0A310SC60 A0A3B5A790 K7J882 A0A158NYY3 A0A1A9V2E7 Q59G70 A0A3B0J032 A0A026W650 B4LIP0 A0A3P9N5Y9 P26572 A0A1I8P9G2 A0A091GC10 D2I5F9 B3KQ18
PDB
1FOA
E-value=1.15443e-112,
Score=1040
Ontologies
PATHWAY
GO
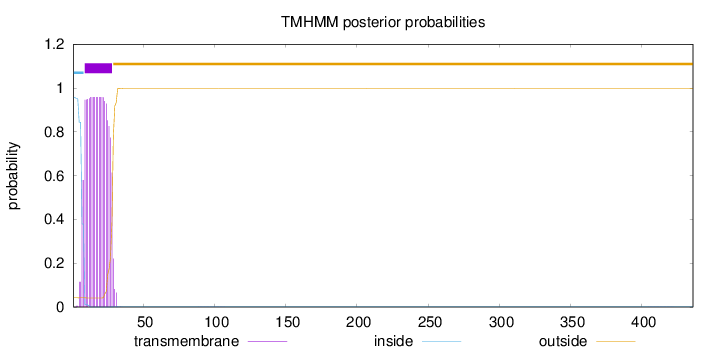
Topology
Subcellular location
Golgi apparatus membrane
Length:
436
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.09929
Exp number, first 60 AAs:
20.0915
Total prob of N-in:
0.95798
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 28
outside
29 - 436
Population Genetic Test Statistics
Pi
264.191679
Theta
167.728786
Tajima's D
3.020227
CLR
0.106247
CSRT
0.978651067446628
Interpretation
Uncertain
